The coronavirus 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a beta-coronavirus that is thought to have been transmitted from bats to humans via an intermediate animal host. Since it originally emerged into the world in December 2019, COVID-19 has infected over 225 million and taken the lives of over 4.6 million worldwide.
 Study: Targeted Isolation of Panels of Diverse Human Broadly Neutralizing Antibodies Against SARS-Like Viruses. Image Credit: ustas7777777 / Shutterstock.com
Study: Targeted Isolation of Panels of Diverse Human Broadly Neutralizing Antibodies Against SARS-Like Viruses. Image Credit: ustas7777777 / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Researchers around the world initially thought that SARS-CoV-2 could be effectively controlled by a safe and effective vaccine, which led to intensive vaccine development efforts. However, while vaccines, especially those based on the messenger ribonucleic acid (mRNA) platform, were largely effective in phase 3 trials, the emergence of variants of concern (VOCs) threatens their efficacy.
Some of these VOCs are much more transmissible than the earlier variants, evade host immunity, or both. The vaccines induced robust neutralizing antibody (nAb) responses; however, with the presence of escape mutations, VOCs have shown resistance to neutralizing immunodominant nAbs elicited by natural infection or vaccination. Thankfully, this loss in neutralizing ability has not translated into a loss of protection against serious disease and death following SARS-CoV-2
With the waning of nAb titers over time, an increasing number of breakthrough infections is being reported. Many scientists fear that this may lead to even more VOCs, as well as serious infections. A few researchers also predict that newer viruses similar to SARS-CoV-2 may cause future pandemics.
Taken together, these observations have led to an exploration of nAbs and vaccines that have broad protective activity against Sarbecoviruses. Convalescent plasma has shown several bnAbs to be present, while scientists have engineered strain-specific nAbs to increase the breadth of neutralization against more SARS-CoV-2 variants.
The aim of such research is to characterize a range of bnAbs to understand what drives breadth of neutralization and to better design immunogens for vaccine development. Many different nAbs have been identified in SARS-CoV-2 infections followed by vaccination with an mRNA vaccine.
Study findings
In the current study, the researchers identified and described 40 bnAbs from two convalescent sera taken from two COVID-19 donors (CC25 and CC84) who later obtained vaccination with mRNA vaccines. These individuals showed the highest levels of nAbs, as well as the highest levels of nAbs with neutralizing activity against multiple Sarbecoviruses that engaged the human angiotensin-converting enzyme 2 (ACE2) receptor. These include SARS-CoV-1 and WIV1, as well as the B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.V2 (Delta) VOCs of SARS-CoV-2.
As expected from earlier research, the infected-vaccinated sera showed comparable efficacy in neutralizing the ancestral SARS-CoV-2 strain and its VOCs, with lower but still enhanced efficacy against SARS-CoV-1. The bnAbs were found to target more conserved spike epitopes.
“SARS-CoV-1/2 cross-neutralization appears to be a good indicator of the presence of pan-sarbecovirus activity and possibly greater CoV neutralization breadth.”
Germline gene enrichment
The two serum samples in the current study yielded over a hundred monoclonal antibodies (mAbs) with cross-reactive SARS-CoV-2/SARS-CoV-2 spike antigens. Seasonal endemic coronavirus specificity was weak. About a third of the mAbs reacted with all the 12 receptor binding domains (RBDs) from all the four clades of sarbecoviruses.
Two-thirds of the mAbs cross-neutralized both SARS-CoV-2 and SARS-CoV-2, while approximately 40% of mAbs neutralized all four sarbecovirus clades.
The germline genes enriched in this collection of mAbs included IGHV3-30, GHV1-46, and IGHV1-69. However, the extent of nucleotide somatic hypermutation in the variable genes did not appear to correlate with the breadth of neutralization. The same lack of correlation was observed with binding to the SARS-CoV-2 RBD.
Over 70% of mAbs which had 20-residue heavy-chain second complementarity determining regions (CDRH2s) used the IGHD2-15 D-gene. Comparably, for 21-residue CDRH3s, either IGHD2-15 or IGHD3-22 germline D-genes were used.
Cross-neutralization by mAbs
With a smaller panel of mAbs, the researchers found that several of these antibodies bound at high affinity to all four sarbecovirus clade RBDs. Of the 30 mAbs examined here, 22 neutralized SARS-CoV-2, whereas 28 were able to neutralize SARS-CoV-1. The latter contained all 22 mAbs of the former category.
Most of the mAbs neutralized SARS-CoV-1 more strongly than SARS-CoV-2, whereas all neutralized WIV1 and most neutralized Pang17. Just a little under half of the mABs showed cross-neutralization against all ACE2-binding Sarbecoviruses. Notably, the three most potent mAbs showed a half-maximal inhibitory concentration (IC50) of 0.04 micrograms (μg)/ milliliter (ml) or less.
Using a few selected bnAbs, the scientists tested for neutralization of the SARS-CoV-2 VOCs. This showed that as the serum samples, the bnAbs were consistently effective against the VOCs, unlike the strain-specific nAb CC12.1. This indicates that the bnAbs recognize conserved RBD epitopes that remain intact, despite escape mutations in the VOCs.
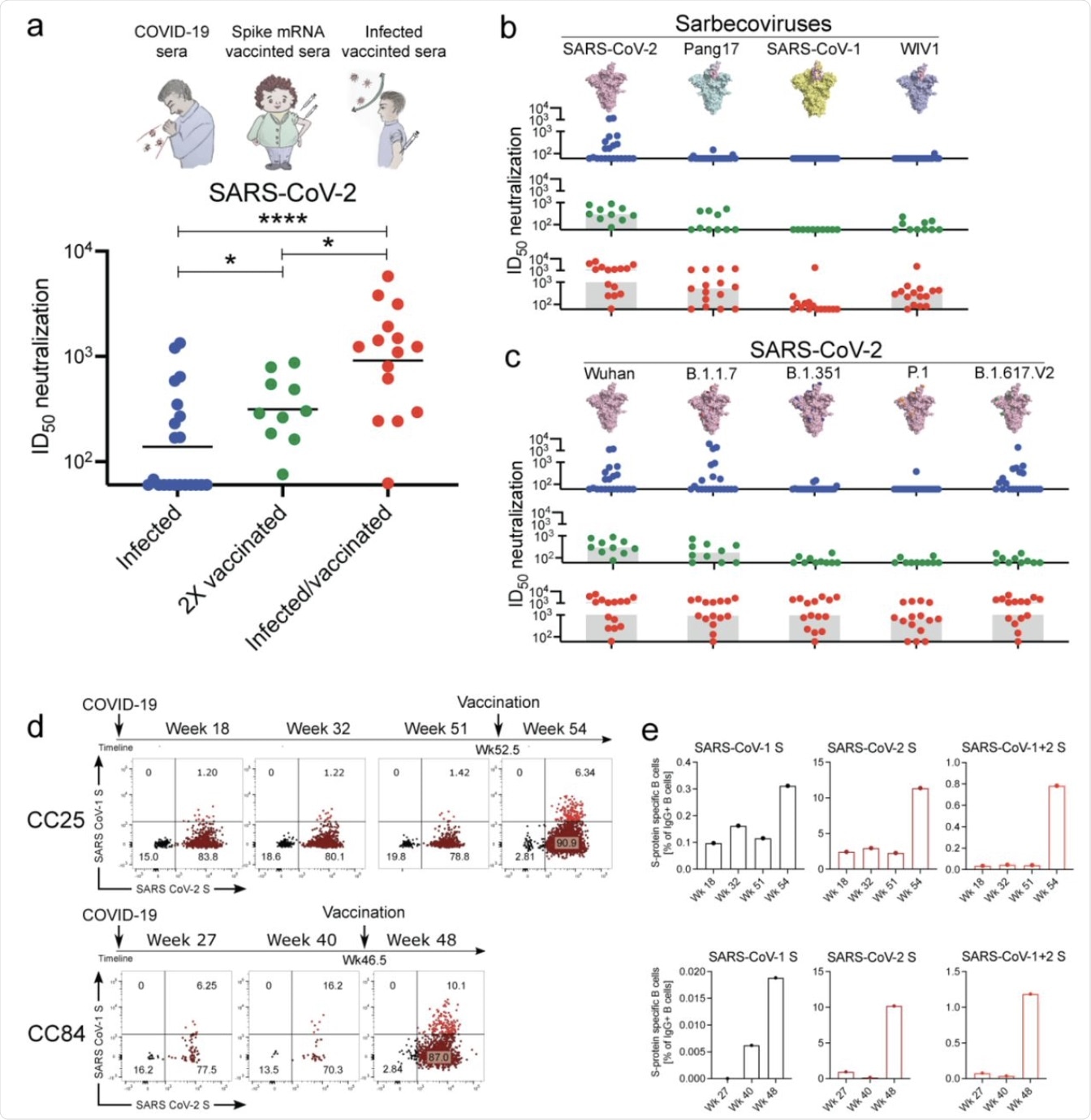 Binding and neutralization of mAbs in terms of potency and breadth.
Binding and neutralization of mAbs in terms of potency and breadth.
A total of 19 mAbs from donor CC25 and 11 mAbs from donor CC84 were selected to determine specificity, neutralization potency and breadth, kinetic properties, and neutralization activities against sarbecoviruses and SARS-CoV-2 VOCs.
a. Heatmap of binding responses determined by BLI using SARS-CoV-1 and SARS-CoV-2 S and S-protein domains and subdomains with IGHV gene usage for each mAb indicated.
b. Heatmap of dissociation constants (KD (M) values) for mAb binding to spike-derived monomeric RBDs from four sarbecovirus clades: clade 1b (n = 3); clade 1a (n = 3); clade 2 (n = 4); clade 3 (n = 2).
c. IC50 neutralization of mAbs against SARS-CoV-2, SARS-CoV-1, Pang17, WIV1, and SHC014 determined using pseudotyped viruses.
d, Neutralization of 18 bnAbs against SARS-CoV-2 (WT) and four major SARS-CoV-2 variants of concern. CC12.1, DH1047, K398.22 and DEN mAbs were used as controls.
Implication
The researchers summed up their findings from “the largest set of human sarbecovirus bnAbs to date,” encoded by a range of immunoglobulin germline gene families, as demonstrating enrichment for specific germline genes. This could indicate the potential value of using these segments to create pan-sarbecovirus vaccines.
“We have identified multiple potent sarbecoviruses bnAbs that exhibit broad reactivity to diverse sarbecovirus lineages.”
The researchers identified several epitopes that were bound by the different groups of bnAbs, with some extending over the RBD and others covering other parts of the viral spike protein. The small epitope variations may be useful in overcoming antibody escape mutations.
Since low-breadth virus-specific nAbs like CC12.1 compete with the more potent bnAbs, the immune response to SARS-CoV-2 VOCs may be impaired. Thus, vaccine designers may consider hiding the epitopes that elicit such specific antibodies, allowing the bnAb sites to remain accessible. Another such approach is engineering the RBD immunogens in the vaccines to elicit bnAbs.
“In particular, as variants emerge during this and future CoV pandemics, the availability of a selection of potent bnAbs provides choice of optimal reagents for prophylaxis and therapy to respond to the viral threats.”
Finally, the combination of infection with vaccination provided a strong bnAb response. This may indicate the presence of small but important differences in the spike protein on native viral particles, as well as the stabilized spike protein used in vaccines. Taken together, the incorporation of these differences into a new vaccine could increase the breadth of the neutralizing antibody response after the combination of infection and vaccination.
The period that elapses between the two exposures may also promote the accumulation of important mutations that elicit bnAbs, such as on intact viral particles within follicular dendritic cells in the lymph node germinal centers. The T-cell immune response to SARS-CoV-2 following infection is possibly better in its robust long-term protection than the antibody response to the vaccine alone.
“Overall, there is an intriguing possibility that pan-sarbecovirus nAb activity may be best achieved by a hybrid approach to immunization that seeks to mimic infection-vaccination, once the key contributing factors to breadth development in that approach can be determined.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
He, W., Musharrafieh, R., Song, G., et al. (2021). Targeted Isolation of Panels of Diverse Human Broadly Neutralizing Antibodies Against SARS-Like Viruses. bioRxiv. doi:10.1101/2021.09.08.459480. https://www.biorxiv.org/content/10.1101/2021.09.08.459480v1
- Peer reviewed and published scientific report.
He, Wan-ting, Rami Musharrafieh, Ge Song, Katharina Dueker, Longping V. Tse, David R. Martinez, Alexandra Schäfer, et al. 2022. “Targeted Isolation of Diverse Human Protective Broadly Neutralizing Antibodies against SARS-like Viruses.” Nature Immunology 23 (6): 960–70. https://doi.org/10.1038/s41590-022-01222-1. https://www.nature.com/articles/s41590-022-01222-1.