In the case of nucleic acid tests, polymerase chain reaction (PCR) has been the gold standard for single-molecule diagnosis. Single-molecule detection of pathogens, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is key to the fight against infectious diseases outbreaks and pandemics.
Currently, simple readouts are obtained using calorimetric sensing with loop-mediated isothermal amplification (LAMP). A new study published in the medRxiv* preprint server finds that plasmonic sensing of LAMP amplicons through deoxyribonucleic acid (DNA) hybridization enables highly specific and single-molecule detection of SARS-CoV-2 RNA.
 Study: Single-Molecule Detection of SARS-CoV-2 by Plasmonic Sensing of Isothermally Amplified Nucleic Acids.
Study: Single-Molecule Detection of SARS-CoV-2 by Plasmonic Sensing of Isothermally Amplified Nucleic Acids.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
PCR tests involve time-consuming protocols and require laboratory infrastructure, which precludes their use in point-of-care (POC) testing. LAMP and other isothermal amplification methods have emerged as alternatives to PCR and allow POC testing.
One problem associated with LAMP is that cannot distinguish template versus non-templated amplification and, as a result, is susceptible to false-positive results. Therefore, it is important to devise novel techniques to directly identify the amplified sequences through a simplified detection scheme. This type of technique will enable the provision of diagnostic platforms with enhanced features like simple readouts, high specificity, and single-molecule detection sensitivity.
About the study
In this new study, scientists have reported a robust method for nucleic acid detection. This method is based on plasmonic sensing of LAMP amplicons through DNA hybridization, termed plasmonic LAMP.
A SARS-CoV-2 RNA model was used for this analysis, wherein researchers showed that the plasmonic LAMP method achieved single-molecule detection which is a useful toolkit to reduce the severity of the ongoing pandemic.
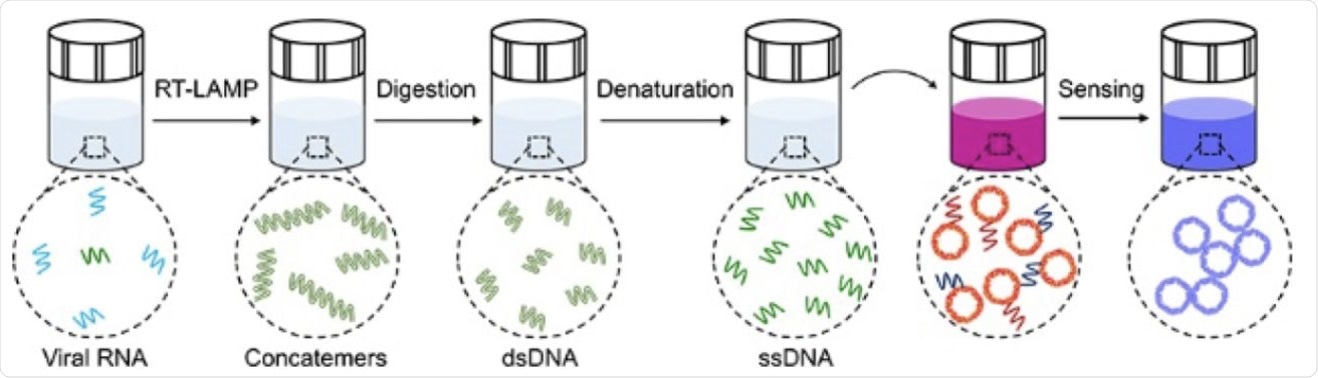
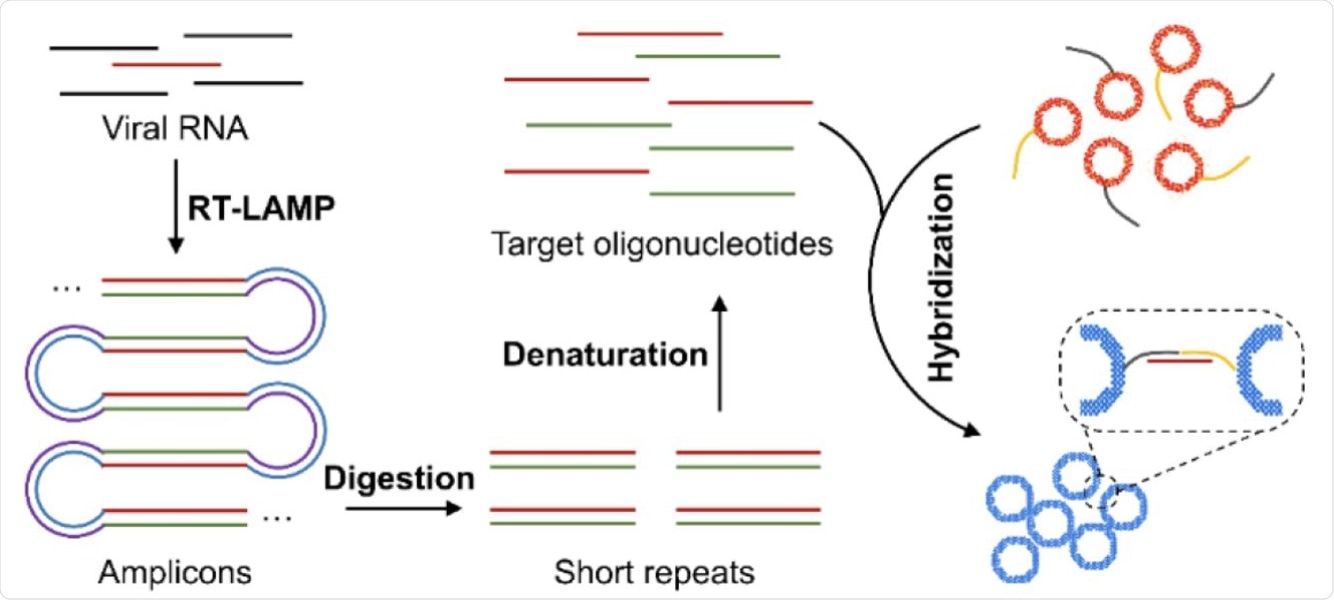 Schematic illustration of the major steps for the proposed plasmonic LAMP single-molecule RNA detection. The viral RNA is first reverse transcribed and amplified into amplicons that are subjected to restriction enzymes digestion, forming short repeats that can be denatured into oligonucleotides for subsequent DNA hybridization linked with plasmonic sensors.
Schematic illustration of the major steps for the proposed plasmonic LAMP single-molecule RNA detection. The viral RNA is first reverse transcribed and amplified into amplicons that are subjected to restriction enzymes digestion, forming short repeats that can be denatured into oligonucleotides for subsequent DNA hybridization linked with plasmonic sensors.
The current study has two main advantages. First, researchers developed gold and silver alloy (Au-Ag) nanoshells, which have four times stronger extinction in the visible wavelengths. Additionally, these nanoshells provide a twenty times lower detection limit for oligonucleotides than Au nanoparticles.
Second, the researchers demonstrated that the new method would enable cutting the complex LAMP amplicons into short repeats. These repeats are amendable for hybridization with oligonucleotide-functionalized nanoshells.
The researchers started with the synthesis of Au-Ag nanoshells by titrating silver nanoparticles with HAuCl4. At a HAuCl4 load of 3.3 mL, the absence of Na3CA led to the formation of thin and porous cages. This could due to the reaction stoichiometry, where one Ag atom is replaced by one Au atom in the case of AuCl2-.
Energy-dispersive X-ray (EDX) mapping of an Au-Ag shell showed that the Au and Ag elements were distributed across the entire particle, thereby, confirming the alloyed structure.
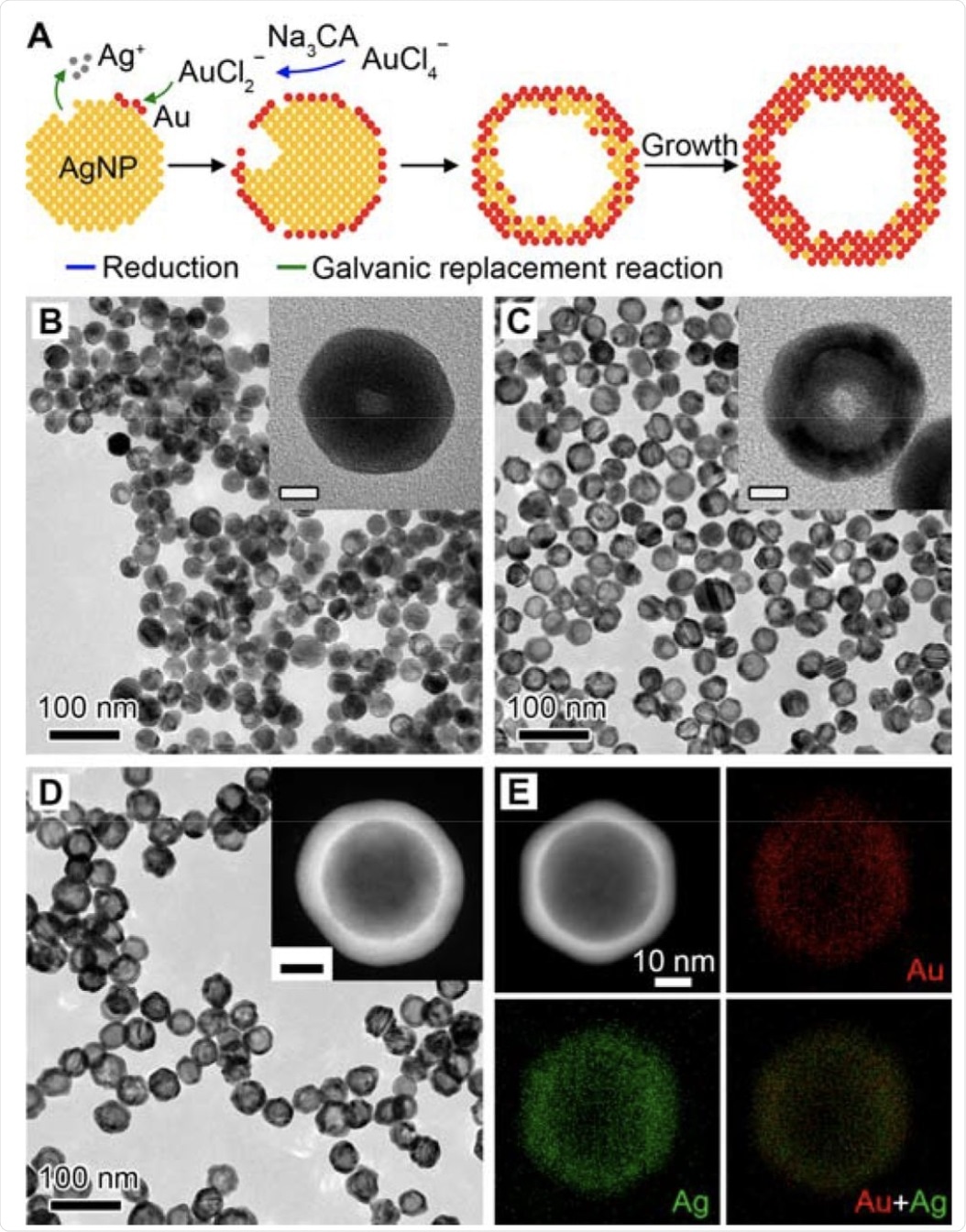 Synthesis and characterization of Au-Ag nanoshells. A Schematic illustration of the Au-Ag shells growth based on galvanic replacement reaction in the presence of Na3CA. B-D TEM images of aliquots taken from the reaction when 1.0 mL (B), 2.5 mL (C), and 10.0 mL (D) HAuCl4 was injected. Insets show the magnified TEM images (B, C) and HAADF-STEM image (D) of individual particles. Scale bars in insets are 10 nm. E EDX mapping image of an individual particle shown in (D).
Synthesis and characterization of Au-Ag nanoshells. A Schematic illustration of the Au-Ag shells growth based on galvanic replacement reaction in the presence of Na3CA. B-D TEM images of aliquots taken from the reaction when 1.0 mL (B), 2.5 mL (C), and 10.0 mL (D) HAuCl4 was injected. Insets show the magnified TEM images (B, C) and HAADF-STEM image (D) of individual particles. Scale bars in insets are 10 nm. E EDX mapping image of an individual particle shown in (D).
The researchers then evaluated the role of Na3CA in shell growth by increasing its concentration. At low concentrations, Na3CA induces a mixture of cages and shells, whereas, in higher concentrations, it yields shells as the final product. The increased deposition rate of Au atoms is essential for the preferential overgrowth of branched or porous shells.
Subsequently, the LSPR properties of Au-Ag nanoshells were investigated, as they are crucial for plasmonic sensing. The extinction spectra clearly revealed a different peak shift between cages and Au-Ag shells.
The extinction peak of Au-Ag shells shifted from 392 nanometers (nm) to 530 nm, while that of cages shifted to longer near-infrared wavelengths. The extinction intensity change of the two structures differed with a monotonic decrease and falling-rising trend, respectively, thus suggesting that shells had higher extinction than the cages. The scientists then performed additional numerical simulations to gain further insights into the LSPR properties of the Au-Ag shells.
The next step was to examine the Au-Ag nanoshells as labels for oligonucleotide detection. A sequence associated with the SARS-CoV-2 N gene was chosen as the target and two complementary sequences were subsequently designed as probes.
Subsequently, the LSPR extinction spectra were recorded and normalized at 537 nm. Because the Au-Ag shells and gold nanoparticles have similar sizes and shapes, the improved LSPR property is likely what caused the observed sensitivity enhancement. A ratiometric quantification method was adopted which improves analytical performance.
The last step was to devise a protocol to combine reverse transcription LAMP (RT-LAMP) with plasmonic Au-Ag nanoshells for single-molecule detection of pathogens such as SARS-CoV-2.
In an effort to demonstrate the importance of its clinical use, the scientists applied the integrated plasmonic LAMP approach to detect nasal swab samples that were spiked with SARS-COV-2 RNA. The analytical performance was impressive and suggested that the Au-Ag-shells-based sensors worked well and had potential uses in the clinical setting. The researchers opined that the established diagnostic approach should be adopted as a nucleic-acid detection platform. However, the set of primers and probing sequences should be changed.
Conclusion
In this study, scientists developed the plasmonic LAMP approach for the single-molecule detection of SARS-CoV-2 RNA. The current work provides a diagnostic toolkit with simple readouts and ultralow detection limits that could potentially be applied in clinical settings.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ye, H., Nowak, C., Liu, Y., et al. (2021) Single-Molecule Detection of SARS-CoV-2 by Plasmonic Sensing of Isothermally Amplified Nucleic Acids. medRxiv. doi:10.1101/2021.10.05.21264561, https://www.medrxiv.org/content/10.1101/2021.10.05.21264561v1
- Peer reviewed and published scientific report.
Ye, Haihang, Chance Nowak, Yaning Liu, Yi Li, Tingting Zhang, Leonidas Bleris, and Zhenpeng Qin. 2022. “Plasmonic LAMP: Improving the Detection Specificity and Sensitivity for SARS-CoV-2 by Plasmonic Sensing of Isothermally Amplified Nucleic Acids.” Small 18 (12): 2107832. https://doi.org/10.1002/smll.202107832. https://onlinelibrary.wiley.com/doi/10.1002/smll.202107832.