Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) continue to threaten the global response to the coronavirus disease 2019 (COVID-19) pandemic. Interestingly, the spike (S) protein sequence of all VOCs is different and displays different sensitivities to interferon-induced transmembrane protein (IFITM) proteins.
 Study: The P681H mutation in the Spike glycoprotein confers Type I interferon 2 resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. Image Credit: Lightspring / Shutterstock.com
Study: The P681H mutation in the Spike glycoprotein confers Type I interferon 2 resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. Image Credit: Lightspring / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
Wuhan-1 S protein pseudotyped lentiviral particles (PLVs) bearing the S protein of each variant were tested to ascertain whether they were restricted by IFITMs overexpressed in A549-ACE2 cells. This confirmed that the D614G mutation in the Wuhan-1 S protein did not affect IFITM1 and IFITM2 sensitivity of PLV entry, while it was slightly enhanced in the presence of IFITM3.
It is worth noting that the Alpha variant, which was primarily investigated in this study to determine the mechanism of IFITM resistance, first appeared in the United Kingdom and quickly became the dominant circulating strain in Europe and North America in early 2021. The researchers found that the Alpha S protein appeared completely insensitive to IFITMs 1, 2, or 3 while the Beta, Gamma, and Delta S proteins still retained some sensitivity to IFITMs 1 and 2.
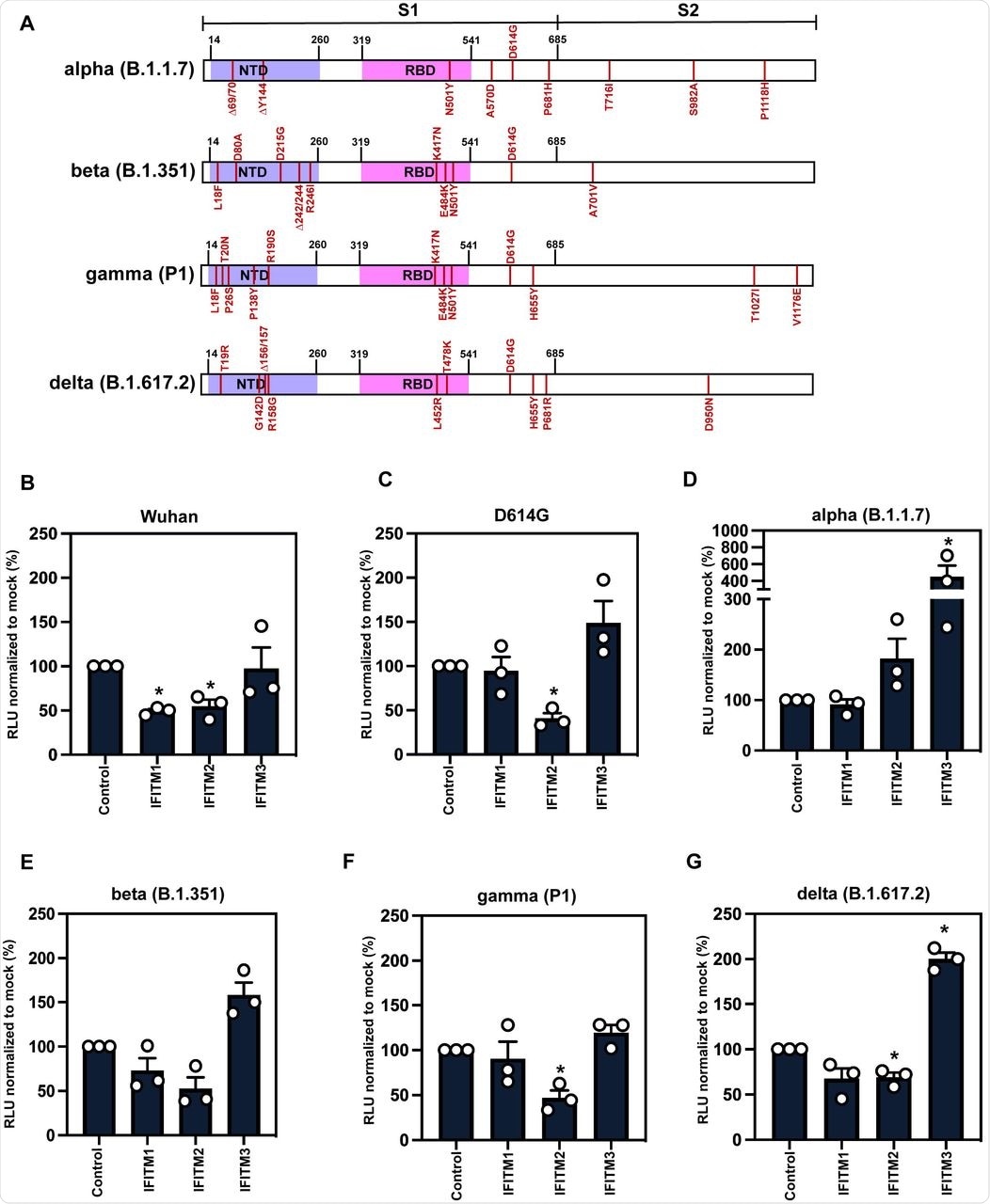 A) Schematic of Spike protein domains of the different variants of concern relative to the original Wuhan Spike sequence: alpha, beta, gamma and delta. The different mutations between the variants are represented in red. B-G) IFITM sensitivity of Wuhan, D614G, alpha, beta, gamma and delta PLVs in A549-ACE2 cells stably expressing the individual IFITMs. PLV entry was quantified by Luciferase activity 48 hours after infection and normalized to control cells. Data shown are mean ± SEM, n=3. Statistics were calculated in Prism using t-test, stars indiciate significance between control cell and individual IFITM (*P=0.05).
A) Schematic of Spike protein domains of the different variants of concern relative to the original Wuhan Spike sequence: alpha, beta, gamma and delta. The different mutations between the variants are represented in red. B-G) IFITM sensitivity of Wuhan, D614G, alpha, beta, gamma and delta PLVs in A549-ACE2 cells stably expressing the individual IFITMs. PLV entry was quantified by Luciferase activity 48 hours after infection and normalized to control cells. Data shown are mean ± SEM, n=3. Statistics were calculated in Prism using t-test, stars indiciate significance between control cell and individual IFITM (*P=0.05).
To confirm the results obtained with the Alpha variant and reiterate them with the full-length virus, researchers infected A549-ACE2 cells with the England/02 isolate and measured infection in these cells after 48 hours. Replication of the isolate was significantly reduced in IFITM2-expressing cells; however, the Alpha variant replicated well in IFITM2-expressing cells and to a significantly higher level in IFITM3-expressing cells compared to the A549-ACE2 cells. This confirmed that the Alpha variant is resistant to IFITMs 1 and 2, whereas IFITM3 enhances its entry.
Previous studies have shown that the Alpha S protein is more efficiently cleaved than Wuhan-1 S protein, which is due to the optimal accessibility of the furin cleavage site (FCS) in the presence of the P681H mutation. Immunoblotting of viral stocks of the England-02 and Alpha variants showed a more processed S protein on the Alpha virions.
PLV infection in E64D-treated A549-ACE2 cells inhibited SARS-CoV-2 entry in endosomal compartments, as E64D is a cathepsin inhibitor that blocks viral entry by cleaving S1/S2 junctions that have not been processed by furin. This suggests that the Alpha S protein-mediated entry of PLVs was found to be significantly less sensitive to the endosomal protease inhibition as compared to the Wuhan-1 S protein, thereby indicating differences in the virus entry site.
Alpha variant is less sensitive to IFNβ than the Wuhan-1 variant
Previous studies have shown that the Wuhan-1 strain of SARS-CoV-2 is highly sensitive to exogenous interferon (IFN) treatment in culture. Since the Alpha variant appears to be resistant to IFITMs expression in A549-ACE2 cells, it was tested with increasing doses of IFNβ.
The observations show that the Alpha variant was more resistant than the Wuhan-like England/02, as indicated by the quantities of viral RNA in A549-ACE2 cells and the human lung epithelial cell line Calu3 48 hours following infection.
Alpha variant displays resistance to type I IFN
To establish the ability of the P681H mutation to confer IFITM and IFNβ resistance to the Alpha variant, the researchers made a recombinant molecular clone of SARS-CoV-2 that mimicked the resistance of the Alpha variant to IFNβ in comparison to the Wuhan-like virus, England-02. This experiment demonstrated that the Alpha S protein is sufficient to bestow type I IFN resistance in A549-ACE2 cells.
When the amino acid residue H681 was reverted to proline in the recombinant virus, this single point mutation conferred a significant sensitivity to IFNβ in Calu3 cells. Lastly, when the researchers knocked down IFITM2 expression using small inhibitory ribonucleic acid (siRNA) in A549-ACE2 cells and treated them with IFNβ, the Alpha variant remained unaffected. This confirmed that the Alpha S protein determines type-I IFN resistance due to the P681H mutation.
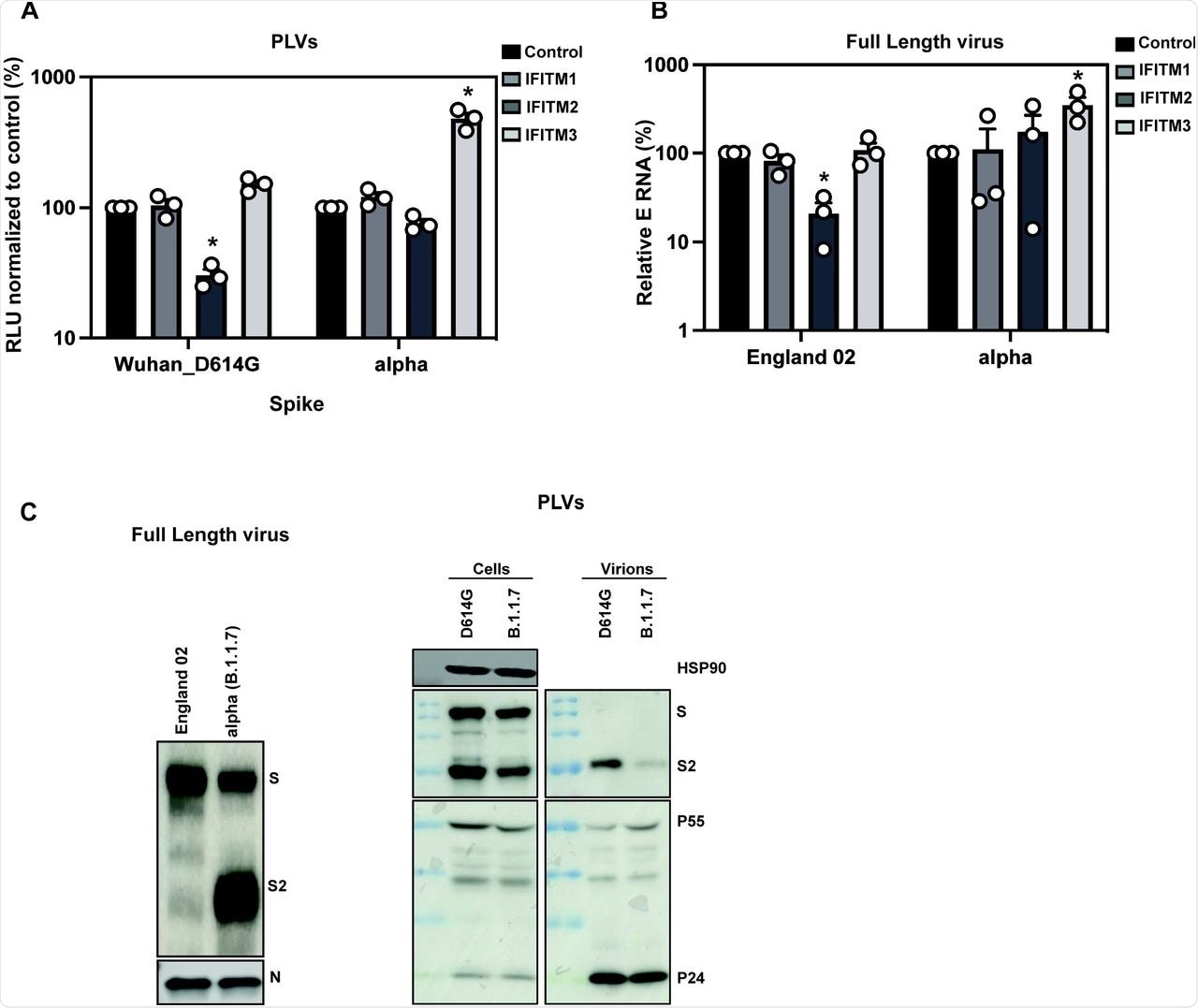 A) D614G and Alpha PLVs infection of A549-ACE2 cells stably expressing the individual IFITMs. Infection was quantified by Luciferase activity 48 hours later and normalized to control cells. Data shown are mean ± SEM, n=3. Statistics were calculated in Prism using t-test, stars indiciate significance between control cell and individual IFITM (*P=<0.05). B) Infection of A549-ACE2 stably expressing the individual IFITMs with England 02 and alpha full-length viruses at MOI 0.01. Infection was quantified by RT-qPCR of E gene relative to GAPDH 48 hours later; graph represents E mRNA levels relative to GAPDH. Data shown are mean ± SEM, n=3. Statistics were calculated in Prism using t-test, stars indiciate significance between control cell and individual IFITM (*P=<0.05). C) Western blot from representative D614G and alpha PLVs produced in HEK293T/17 cells, and virions from full-length England-02 and alpha viruses. Virions were purified through a 20% sucrose gradient.
A) D614G and Alpha PLVs infection of A549-ACE2 cells stably expressing the individual IFITMs. Infection was quantified by Luciferase activity 48 hours later and normalized to control cells. Data shown are mean ± SEM, n=3. Statistics were calculated in Prism using t-test, stars indiciate significance between control cell and individual IFITM (*P=<0.05). B) Infection of A549-ACE2 stably expressing the individual IFITMs with England 02 and alpha full-length viruses at MOI 0.01. Infection was quantified by RT-qPCR of E gene relative to GAPDH 48 hours later; graph represents E mRNA levels relative to GAPDH. Data shown are mean ± SEM, n=3. Statistics were calculated in Prism using t-test, stars indiciate significance between control cell and individual IFITM (*P=<0.05). C) Western blot from representative D614G and alpha PLVs produced in HEK293T/17 cells, and virions from full-length England-02 and alpha viruses. Virions were purified through a 20% sucrose gradient.
Mutations in SARS-CoV-2 VOCs help them replicate faster and adapt to resist innate immunity
To summarize, the Alpha S protein has undergone several mutations in the polybasic cleavage site that enhance S protein cleavage, including the P681H mutation. The Alpha S protein confers a level of resistance to the effects of IFNβ in A549-ACE2 cells.
This co-relates with resistance to restriction mediated by IFITM2 and a marked infection enhancement by IFITM3. Furthermore, the P681H mutation is mandatory for comparative resistance to IFNβ in a recombinant molecular clone of SARS-CoV-2 encoding the Alpha S protein.
Taken together, the findings suggest that in addition to adaptive immune escape, mutations in VOCs help them replicate faster and adapt to resist innate immunity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lista, M. J., Winstone, H., Wilson, H. D., et al. (2021). The P681H mutation in the Spike glycoprotein confers Type I interferon 2 resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. bioRxiv. doi:10.1101/2021.11.09.467693. https://www.biorxiv.org/content/10.1101/2021.11.09.467693v1.
- Peer reviewed and published scientific report.
Lista, Maria Jose, Helena Winstone, Harry D. Wilson, Adam Dyer, Suzanne Pickering, Rui Pedro Galao, Giuditta De Lorenzo, et al. 2022. “The P681H Mutation in the Spike Glycoprotein of the Alpha Variant of SARS-CoV-2 Escapes IFITM Restriction and Is Necessary for Type I Interferon Resistance.” Edited by Stacey Schultz-Cherry. Journal of Virology, November. https://doi.org/10.1128/jvi.01250-22. https://journals.asm.org/doi/10.1128/jvi.01250-22.