People affected by coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) primarily suffer from respiratory illness. However, COVID-19 has also been reported to cause neurological symptoms in certain sections of the affected population.
Neuroimaging and cognitive screening have identified that COVID-19 induces impairment of the frontal cortex, which plays a vital role in cognitive function. It has also been suggested that COVID-19 results in long-term cognitive impairment. However, the molecular changes induced by COVID-19 that lead to cognitive impairment are yet to be investigated.
Natural aging results in decreased frontal cortex activity that may lead to a cognitive deficit. The molecular signatures associated with aging in the human brain are increased immune signaling and decreased synaptic activity.
As COVID-19 has been found to cause frontal cortex impairment similar to aging, the scientists in a recent study attempted to investigate if severe COVID-19 resulted in aging-related molecular signatures in the brain. The findings from this study are published as a preprint on the medRxiv* server.
 Study: Severe COVID-19 induces molecular signatures of aging in the human brain. Image Credit: Kateryna Kon / Shutterstock
Study: Severe COVID-19 induces molecular signatures of aging in the human brain. Image Credit: Kateryna Kon / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Severe COVID-19 infection causes distinct global transcriptomic changes in the frontal cortex
In the study, whole transcriptomic analyses were performed on the postmortem frontal cortex of twelve COVID-19 patients compared with twelve age-matched and sex-matched uninfected controls.
Clustering analysis via t-distributed stochastic neighbor embedding (TSNE) was performed, which indicated that the transcriptomic profiles of COVID-19 patients were distinct from the controls. Interestingly, the transcriptomic profiles of two of the controls were aged 71 and 84 were found to be similar to that of the COVID-19 patients.
Quantitative PCR (qPCR) analysis showed the absence of SARS-CoV-2 in the frontal cortex of both the COVID-19 patients and controls at the time of death. This indicates that the gene expression changes observed in COVID-19 patients were not due to the direct action of the virus on the frontal cortex.
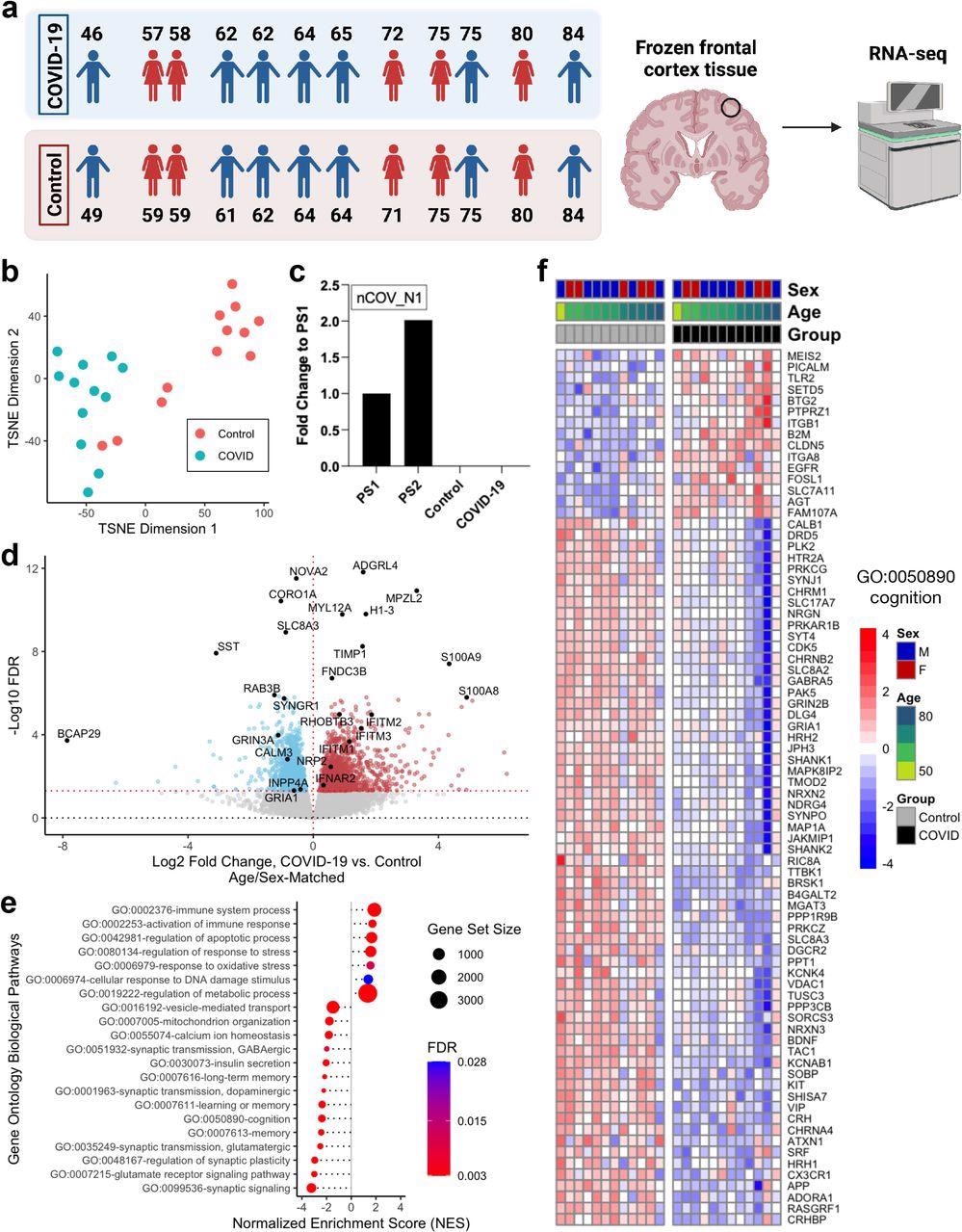 Severe COVID-19 induces global transcriptomic changes in the frontal cortex of the brain. a. Left, age and sex of each individual in COVID-19 or control groups (n=12/group) analyzed in this cohort. Each COVID-19 case was matched with an uninfected control case by sex and age (±2 years). Right, schematic of the study approach. Schematic was generated with BioRender. b. t-distributed stochastic neighbor embedding (TSNE) analysis of frontal cortex transcriptomes from COVID-19 cases and uninfected controls. c. qPCR assessment of SARS-CoV-2 viral RNA in the frontal cortex using the nCOV_N1 primer set. PS1 and PS2 correspond to the 2019-nCoV_N_Positive Control RUO Plasmid (IDT) at concentrations of 1,000 and 2,000 copies/μl, respectively (a technical duplicate/concentration was used to estimate the corresponding mean; for control and COVID-19 samples n=13/group). d. Volcano plot representing the differentially expressed genes (DEGs) of the frontal cortex of COVID-19 cases versus matched controls. Red points, significantly upregulated genes among COVID-19 cases (false discovery rate < 0.05). Blue points, significantly downregulated genes among COVID-19 cases. Black points, highlighted significant genes with corresponding gene symbols. e. Gene ontology (GO) biological pathway enrichment analysis of COVID-19 versus control brain DEGs. Gene ranks were determined by signed -log10 false discovery rates of DEGs. FDR, gene set enrichment analysis false discovery rate. f. Heatmap of relative gene expression levels of significant DEGs associated with the “cognition” (GO: 0050890) GO term across COVID-19 and control samples. Color legend, scaled gene expression levels across subjects, normalized via variance-stabilized transformation.
Severe COVID-19 induces global transcriptomic changes in the frontal cortex of the brain. a. Left, age and sex of each individual in COVID-19 or control groups (n=12/group) analyzed in this cohort. Each COVID-19 case was matched with an uninfected control case by sex and age (±2 years). Right, schematic of the study approach. Schematic was generated with BioRender. b. t-distributed stochastic neighbor embedding (TSNE) analysis of frontal cortex transcriptomes from COVID-19 cases and uninfected controls. c. qPCR assessment of SARS-CoV-2 viral RNA in the frontal cortex using the nCOV_N1 primer set. PS1 and PS2 correspond to the 2019-nCoV_N_Positive Control RUO Plasmid (IDT) at concentrations of 1,000 and 2,000 copies/μl, respectively (a technical duplicate/concentration was used to estimate the corresponding mean; for control and COVID-19 samples n=13/group). d. Volcano plot representing the differentially expressed genes (DEGs) of the frontal cortex of COVID-19 cases versus matched controls. Red points, significantly upregulated genes among COVID-19 cases (false discovery rate < 0.05). Blue points, significantly downregulated genes among COVID-19 cases. Black points, highlighted significant genes with corresponding gene symbols. e. Gene ontology (GO) biological pathway enrichment analysis of COVID-19 versus control brain DEGs. Gene ranks were determined by signed -log10 false discovery rates of DEGs. FDR, gene set enrichment analysis false discovery rate. f. Heatmap of relative gene expression levels of significant DEGs associated with the “cognition” (GO: 0050890) GO term across COVID-19 and control samples. Color legend, scaled gene expression levels across subjects, normalized via variance-stabilized transformation.
Further, the scientists identified 2,809 differentially expressed genes (DEGs) bearing unique Ensembl gene IDs when the COVID-19 transcriptomic profiles were compared to age-matched and sex-matched controls. Amongst the DEGs, 1,397 were found to be upregulated, and 1,412 were downregulated.
Circulating calprotectin levels is a biomarker that distinguishes severe COVID-19 from mild disease and in the present study, S100A8/S100A9 genes that encode calprotectin were upregulated in COVID-19 patient cohort.
Further, similar to earlier reports in COVID-19 patients, SYNGR1 levels were downregulated in the patient cohort. The gene expression changes in COVID-19 patients observed in this study correlate with evidence from previous reports.
![Overview of differential expression patterns in frontal cortex of COVID-19 and uninfected subjects. a. Age and sex matching used for differential expression analysis. b. Heatmap of relative gene expression levels of all significant DEGs (FDR < 0.05) across COVID-19 and control samples. Color legend, scaled gene expression levels across subjects, normalized via variance-stabilized transformation. c. Heatmap of relative expression levels of genes previously implicated in SARS-CoV-2 viral entry in the human brain [8] across COVID-19 and control samples. Color legend, scaled gene expression levels across patients, normalized via variance-stabilized transformation. Light gray cells, no aligned reads or genes filtered out of analysis. Overview of differential expression patterns in frontal cortex of COVID-19 and uninfected subjects. a. Age and sex matching used for differential expression analysis. b. Heatmap of relative gene expression levels of all significant DEGs (FDR < 0.05) across COVID-19 and control samples. Color legend, scaled gene expression levels across subjects, normalized via variance-stabilized transformation. c. Heatmap of relative expression levels of genes previously implicated in SARS-CoV-2 viral entry in the human brain [8] across COVID-19 and control samples. Color legend, scaled gene expression levels across patients, normalized via variance-stabilized transformation. Light gray cells, no aligned reads or genes filtered out of analysis.](https://www.news-medical.net/images/news/ImageForNews_697883_16381604742578654.jpg) Overview of differential expression patterns in the frontal cortex of COVID-19 and uninfected subjects. a. Age and sex matching used for differential expression analysis. b. Heatmap of relative gene expression levels of all significant DEGs (FDR < 0.05) across COVID-19 and control samples. Color legend, scaled gene expression levels across subjects, normalized via variance-stabilized transformation. c. Heatmap of relative expression levels of genes previously implicated in SARS-CoV-2 viral entry in the human brain [8] across COVID-19 and control samples. Color legend, scaled gene expression levels across patients, normalized via variance-stabilized transformation. Light gray cells, no aligned reads or genes filtered out of the analysis.
Overview of differential expression patterns in the frontal cortex of COVID-19 and uninfected subjects. a. Age and sex matching used for differential expression analysis. b. Heatmap of relative gene expression levels of all significant DEGs (FDR < 0.05) across COVID-19 and control samples. Color legend, scaled gene expression levels across subjects, normalized via variance-stabilized transformation. c. Heatmap of relative expression levels of genes previously implicated in SARS-CoV-2 viral entry in the human brain [8] across COVID-19 and control samples. Color legend, scaled gene expression levels across patients, normalized via variance-stabilized transformation. Light gray cells, no aligned reads or genes filtered out of the analysis.
Differentially expressed genes in the frontal cortex of severe COVID-19 patients are similar to those induced by aging
Pathway enrichment analysis was performed using annotated Gene Ontology (GO) Biological processes to establish the functional roles of the observed gene expression changes in the transcriptome. In this analysis, the input gene set is compared with each of the terms or bins in the GO and a statistical test is performed for each term or bin to determine if it is enriched for the input genes.
Interestingly, the scientists observed significant enrichment of several DEGs and GO terms that were related to the aging of the human brain. In the COVID-19 patient cohort, along with positive enrichment of terms associated with immune response, the related genes such as BCL2, IFI16, and CFH were upregulated.
The expression levels of IFITM1-3, associated with interferon response, were highly dysregulated with elevated levels of expression. Synaptic function GO terms including synaptic signaling, regulation of synaptic plasticity, glutamatergic, GABAergic, dopaminergic synaptic transmission were found to be negatively enriched, and this correlated with the observed downregulation of genes involved in synaptic signalings such as SST, GRIA1, and GRIN2B.
Notably, SST is a gene that has previously been reported to be related to aging in the frontal cortex of humans, and it has been observed to be the most downregulated gene in the COVID-19 patient cohort in the present study.
The scientists in the present study observed significant enrichment of GO terms corresponding to the cellular response to DNA damage, mitochondrial function, regulation of response to stress and oxidative stress, vesicular transport, calcium homeostasis, apoptosis, and insulin signaling, and pathways known to be associated with aging and specifically brain aging. Further, GO terms corresponding to cognitive function, memory, and learning were additionally enriched.
The DEGs overlapping with these enriched pathways was assessed, and genes such as the brain-derived neurotrophic factor (BDNF) known to be related to aging were identified.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathway enrichment analyses were performed, and the findings suggest positive enrichment of immune activation and negative enrichment of synaptic function pathways.
Notably, the “Coronavirus Disease - COVID-19” pathway associated with the non-neuronal tissue effects of COVID-19 was identified as a significantly enriched pathway in the COVID-19 patient cohort in the present study.
The findings suggest that changes in biological pathways that are related to natural aging are also observed in COVID-19 patients with severe disease.
Severe COVID-19 induced aging effects are more pronounced in the brains of younger COVID-19 patients
The scientists further investigated if severe COVID-19 induced changes in the transcriptome are similar to the changes induced by aging in the human brain. They assessed transcriptome-wide datasets from five patients for aging-related changes and found that genes that were upregulated or downregulated during aging were similarly upregulated or downregulated in COVID-19 patients with severe disease. Particularly a gene set that was known to be associated with aging was significantly upregulated in the COVID-19 patient cohort in this study.
The findings were further confirmed through quantitative PCR (qPCR) analyses, where genes S100A9, MYL12A, and RHOBTB3 were found to be upregulated and genes CALM3, INPP4A, GRIA1, and GRIN3A were found to be downregulated in the frontal cortex of COVID-19 patients.
Interestingly, the same set of genes were also differentially expressed in the frontal cortex from aged individuals.
The scientists further attempted to study if the aging gene signature differs between the younger COVID-19 patients aged 65 years or below and the older COVID-19 patients aged above 65 years of age.
Interestingly, in young COVID-19 patients, the scientists observed higher levels of changes in gene expression when compared to the older COVID-19 patients.
In the case of younger COVID-19 patients, 1,631 upregulated genes and 2,073 downregulated genes were identified that matched with several DEGs of age/sex-matched controls. However, in older COVID-19 patients, upregulated expression of 19 genes, including HBA1, HBA2, and HBB genes, and downregulated expression of 4 genes were observed.
These DEGs also exhibit the same trends associated with aging-related genes in the frontal cortex. These findings indicate that COVID-19 induced aging effects are more pronounced in the brains of younger COVID-19 patients than older ones.
Further studies with young COVID-19 patient cohorts will confirm the findings observed.
The scientists also investigated if the COVID-19 induced molecular changes differed based on gender. They found that the COVID-19 induced changes in aging-related genes and pathways consistent between male and female COVID-19 patients.
Conclusion
The present study is the first to demonstrate similarities in the transcriptomic profiles of the frontal cortex of COVID-19 patients and the aging human brain.
The findings from the present study suggest that severe COVID-19 disease may result in aging-related changes in the brain and may result in premature aging. These changes are more profound in younger patients compared to older ones. Earlier reports indicate a residual cognitive deficit in recovered COVID-19 patients.
The study further indicates that increased rates of cognitive impairment and neurodegeneration may occur as long-term effects of long COVID. Therefore, it may be necessary to monitor recovered COVID-19 patients for aging related neurological disorders regularly.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Severe COVID-19 induces molecular signatures of aging in the human brain Maria Mavrikaki, Jonathan D. Lee, Isaac H. Solomon, Frank J. Slack medRxiv 2021.11.24.21266779; doi: https://doi.org/10.1101/2021.11.24.21266779, https://www.medrxiv.org/content/10.1101/2021.11.24.21266779v1
- Peer reviewed and published scientific report.
Mavrikaki, Maria, Jonathan D. Lee, Isaac H. Solomon, and Frank J. Slack. 2022. “Severe COVID-19 Is Associated with Molecular Signatures of Aging in the Human Brain.” Nature Aging, December, 1–8. https://doi.org/10.1038/s43587-022-00321-w. https://www.nature.com/articles/s43587-022-00321-w.