The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Mu variant (B.1.621, B.1.621.1) has been tagged as a Variant Being Monitored (VBM), and as of August 30, 2021, it had been detected in 39 countries.
Colombia was identified as the epicenter of SARS-CoV-2 infection caused by the Mu variant. A huge surge of COVID-19 cases was reported in Columbia between March and July 2021, and SARS-CoV-2 infections caused by the gamma variant were found to be prevalent during the early stages of the surge. However, by May, the infections due to the Mu variant became dominant in Columbia.
Reports from WHO suggest that the Mu variant bears mutations that increase the risk of resistance to currently available vaccines and that further investigations are required in this area. The Mu variant is still not considered as a variant of concern by WHO.
The scientists in the present study have earlier demonstrated that the Mu variant exhibits resistance to antibodies produced in response to natural SARS-CoV-2 infection and the anti-SARS-CoV-2 vaccine Pfizer-BioNTech (BNT162b2).
The most common mutations identified in the Mu variant include the T95I and YY144-145TSN mutations in the N-terminal domain, R346K, E484K, and N501Y mutations in the receptor-binding domain, and D614G, P681H, and D950N mutations, which occur in different regions of the spike protein.
The research, released as a preprint on the bioRxiv* server, attempted to answer the cardinal question, “what are the mutations in Mu variants that confer resistance to COVID-19 convalescent sera and vaccine sera?”
 Study: Characterization of the immune resistance of SARS-CoV-2 Mu variant and the immunity induced by Mu infection. Image Credit: Fit Ztudio / Shutterstock
Study: Characterization of the immune resistance of SARS-CoV-2 Mu variant and the immunity induced by Mu infection. Image Credit: Fit Ztudio / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
YY144-145TSN or E484K mutations contribute to the SARS-CoV-2 Mu variant’s resistance to antiviral sera
Plasmids were generated by means of site-directed overlap extension PCR, and they express the SARS-CoV-2 spike protein derivatives of parental D614G (B.1) and Mu (B.1.621) variant.
Neutralization assays were performed with a series of pseudoviruses harboring these plasmids bearing the mutations. The assay was additionally performed employing pseudovirus harboring the plasmids that express spike proteins of the Alpha, Beta, Gamma, Delta, Lambda, or Mu variants.
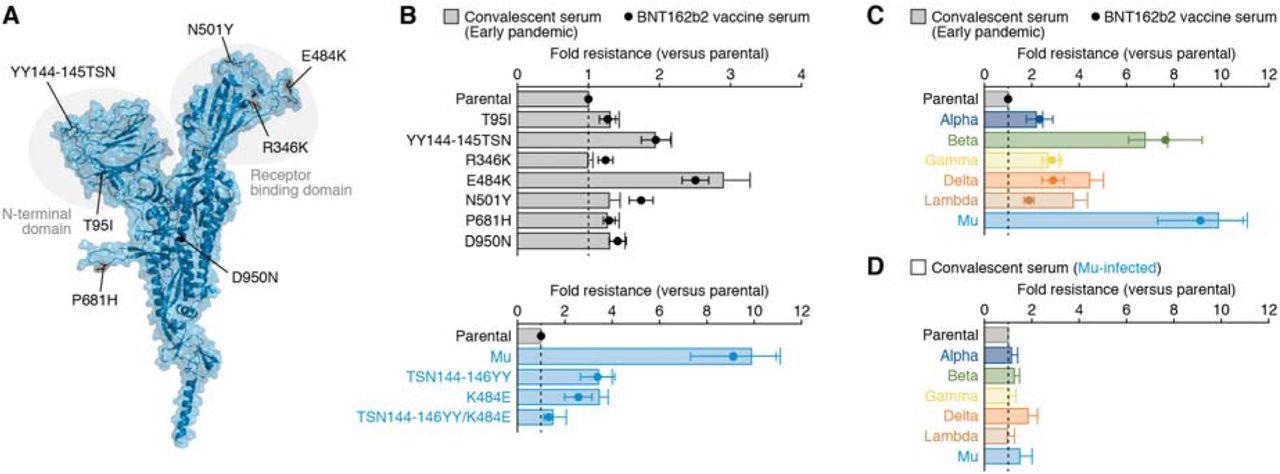 Characterization of the immune resistance of the Mu variant. Panel A shows the position of the mutations in the Mu variant. Cartoon and surface models are overlayed. The mutations in the Mu variant are indicated. The structure of the N-terminal domain is shown in Fig. S1 in the Supplementary Appendix. Panels B to D shows the results of virus neutralization assays. Neutralization assays were performed with the use of pseudoviruses harboring the SARS-CoV-2 spike proteins of parental virus (the B.1 lineage virus, which harbors the D614G mutation)-based derivatives (Panel B, top), the spike proteins of Mu-based derivatives (Panel B, bottom), or the spike proteins of the Alpha, Beta, Gamma, Delta, Lambda or Mu variants (Panels C and D). Serum samples were obtained from 15 convalescent persons who had been infected with SARS-CoV-2 in the early pandemic, 14 persons who had received the BNT162b2 vaccine, and 4 convalescent persons who had been infected with SARS-CoV-2 Mu variant. In Panels B and C, the heights of the bars (serum samples obtained from the convalescent persons who had been infected with SARS-CoV-2 in the early pandemic) and the circles (serum samples obtained from the persons who had received the BNT162b2 vaccine) indicate the average difference in neutralization resistance of the indicated variants as compared with that of the parental virus. In Panel D, the heights of the bars (serum samples obtained from the convalescent persons who had been infected with Mu variant) indicate the average difference in neutralization resistance of the indicated variants as compared with that of the parental virus. The error bars indicate the standard error of the mean. The vertical dashed lines indicate value 1.
Characterization of the immune resistance of the Mu variant. Panel A shows the position of the mutations in the Mu variant. Cartoon and surface models are overlayed. The mutations in the Mu variant are indicated. The structure of the N-terminal domain is shown in Fig. S1 in the Supplementary Appendix. Panels B to D shows the results of virus neutralization assays. Neutralization assays were performed with the use of pseudoviruses harboring the SARS-CoV-2 spike proteins of parental virus (the B.1 lineage virus, which harbors the D614G mutation)-based derivatives (Panel B, top), the spike proteins of Mu-based derivatives (Panel B, bottom), or the spike proteins of the Alpha, Beta, Gamma, Delta, Lambda or Mu variants (Panels C and D). Serum samples were obtained from 15 convalescent persons who had been infected with SARS-CoV-2 in the early pandemic, 14 persons who had received the BNT162b2 vaccine, and 4 convalescent persons who had been infected with SARS-CoV-2 Mu variant. In Panels B and C, the heights of the bars (serum samples obtained from the convalescent persons who had been infected with SARS-CoV-2 in the early pandemic) and the circles (serum samples obtained from the persons who had received the BNT162b2 vaccine) indicate the average difference in neutralization resistance of the indicated variants as compared with that of the parental virus. In Panel D, the heights of the bars (serum samples obtained from the convalescent persons who had been infected with Mu variant) indicate the average difference in neutralization resistance of the indicated variants as compared with that of the parental virus. The error bars indicate the standard error of the mean. The vertical dashed lines indicate value 1.
The neutralization activity of serum samples collected from 15 COVID-19 convalescent individuals who were infected in April 2020 during the early stages of the pandemic was tested in the virus neutralization assays. Additionally, serum samples from 14 subjects who were administered the BNT162b2 vaccine were also tested.
The scientists identified that two mutations YY144-145TSN and E484K, were responsible for the resistance to antibodies elicited by SARS-CoV-2 infection and the anti-SARS-CoV-2 vaccine.
This was further confirmed using ‘loss of function’ experiments on a series of Mu-based pseudoviruses that have lost the mutations of interest. Interestingly, it was found that reverting the YY144-145TSN or E484K mutations in the spike protein of the Mu variant resulted in a loss of neutralization resistance.
These findings suggest that the two mutations YY144-145TSN or E484K are responsible for the resistance exhibited by the Mu variants against the neutralizing antibodies.
Sera from SARS-CoV-2 Mu variant infected individuals exhibit broad-spectrum antiviral activity
Further assessment of the immunological spectrum of serum samples collected from COVID-19 convalescents was conducted in the study. The Mu variant was found to be nine times resistant to the sera collected from vaccinated individuals and convalescents who had natural SARS-CoV-2 infection during the early days of the pandemic.
Notably, the Mu variant did not exhibit resistance to sera collected from individuals infected by the Mu variant. Additionally, the sera from Mu variant infected individuals also showed broad-spectrum neutralization activity against different variants of concern such as alpha, beta, gamma and, lambda.
Conclusion
Emerging variants of SARS-CoV-2 are concerning. They need to be monitored carefully as they show increased transmissibility, pathogenicity, and have a high risk of resistance to the immune response compared to the original strains.
The scientists in the present study have shown that the Mu variant is resistant to neutralizing antibodies in the sera of COVID-19 convalescents and vaccinated individuals. Further, the sera from Mu-infected individuals showed broad-spectrum antiviral activity.
The sequence of the Mu variant can be utilized to develop vaccines that exhibit broad-spectrum antiviral activity against the original SARS-CoV-2 strain and its variants.
Further, research in this area is required to generate Mu variant-based novel vaccine candidates that can provide effective prophylactic therapy for COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Sources:
Journal references:
- Preliminary scientific report.
Uriu, K. et al. Characterization of the immune resistance of SARS-CoV-2 Mu variant and the immunity induced by Mu infection. 2021.11.23.469770, (2021) doi:10.1101/2021.11.23.469770, https://doi.org/10.1101/2021.11.23.469770
- Peer reviewed and published scientific report.
Keiya Uriu, Paúl Cárdenas, Erika Muñoz, Veronica Barragan, Yusuke Kosugi, Kotaro Shirakawa, Akifumi Takaori-Kondo, Ecuador-COVID19 Consortium, The Genotype to Phenotype Japan (G2P-Japan) Consortium , Kei Sato. 2023. "Characterization of the Immune Resistance of Severe Acute Respiratory Syndrome Coronavirus 2 Mu Variant and the Robust Immunity Induced by Mu Infection." The Journal of Infectious Diseases. https://doi.org/10.1093/infdis/jiac053. https://academic.oup.com/jid/article/226/7/1200/6530359.