Since coronavirus 2019 (COVID-19) vaccines have started to be mass-produced, many developed countries have initiated programs designed to immunize as many people as possible. However, many of the new variants such as the Delta and Beta strains have shown the ability to evade vaccine-induced immunity.
Researchers from ImmunityBio, Inc.. the Infectious Disease Research Institute (IDRI), and the University of Washington have been investigating heterologous vaccination (vaccination with two different vaccines) as a potential solution to this problem. The two vaccines are a next-generation human adenovirus serotype 5 (hAd5)-vectored dual-antigen spike (S) and nucleocapsid (N) vaccine (AdS+N) and a self-amplifying and -adjuvanted S RNA vaccine (SASA S) delivered by a nanostructured lipid carrier.
Study: Heterologous Vaccination with SARS-CoV-2 Spike saRNA Prime followed by DNA Dual-Antigen Boost Induces Robust Antibody and T-Cell Immunogenicity against both Wild Type and Delta Spike as well as Nucleocapsid Antigens. Image Credits: Mongkolchon Akesin/Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A preprint version of the group’s study is available on the bioRxiv* server, while the article undergoes peer review.
The study
The researchers used the next-generation vector hAd5 to create viral vaccine constructs. This vector has additional deletions in the E2b region that remove expression of viral DNA polymerase and in preterminal protein genes, preventing propagation in human cell lines. The AdS+N vaccine expresses a spike protein sequence modified with a fusion linker peptide sequence, as well as a nucleocapsid sequence with an enhanced T-cell stimulation domain (ESTD) signal to direct the translated nucleocapsid protein to the endosomal pathway. Targeting both the nucleocapsid and spike proteins reduces the risk that new variants will be able to evade the immune response, as most mutations target the receptor-binding domain (RBD) of the spike protein.
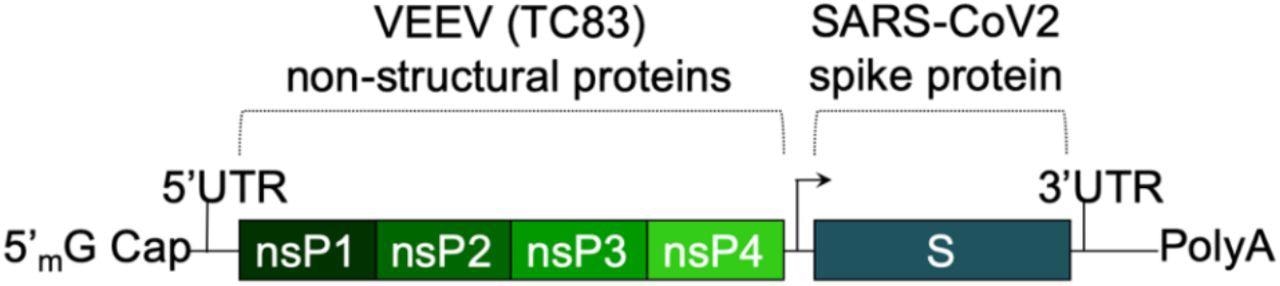 The saRNA(D614G)-2P-3Q-NLC (SASA S) vaccine. The SASA S vaccine comprises an saRNA replicon backbone consisting of the non-structural protein (nsPs) 1-4 derived from the Venezuelan equine encephalitis virus (VEEV) vaccine strain TC-83 and an independent open reading frame under the control of a subgenomic promoter sequence that contains Wuhan sequence S with a diproline (pp) mutation and a QQAQ furin cleavage site sequence.
The saRNA(D614G)-2P-3Q-NLC (SASA S) vaccine. The SASA S vaccine comprises an saRNA replicon backbone consisting of the non-structural protein (nsPs) 1-4 derived from the Venezuelan equine encephalitis virus (VEEV) vaccine strain TC-83 and an independent open reading frame under the control of a subgenomic promoter sequence that contains Wuhan sequence S with a diproline (pp) mutation and a QQAQ furin cleavage site sequence.
The SASA S vaccine uses a saRNA replicon that expresses the spike protein. The spike RNA sequence is codon-optimized and expresses both the wild-type spike protein and the D614G mutation, found in most dominant variants. A prefusion conformation stabilizing diproline mutation is also included, and the RRAR furin cleavage site sequence is replaced with a QQAQ sequence.
The vaccines were administered to mice via subcutaneous injection, and later blood was collected and sera isolated from each mice. Splenocytes were also isolated from the mice. The researchers performed intracellular cytokine stimulation (ICS) on the splenocytes and used ELISpot assays to detect cytokines secreted from the same splenocytes. Enzyme-linked immunosorbent assays (ELISAs) were used for IgG antibody detection in mouse sera, examining spike S1 domain binding, Delta spike S1 domain binding, and nucleocapsid protein binding. The scientists also performed lentiviral pseudovirus neutralization assays on the sera to examine the ability of the sera to neutralize the wild-type, Delta, and Beta strains.
The ELISAs showed that mice receiving the SASA S vaccine in any regimen showed the highest levels of anti-spike protein IgGs. Any mice that received vaccines displaying the nucleocapsid proteins generated anti-nucleocapsid IgGs, as expected, but all showed similar levels of reaction against the nucleocapsid. Against both the Delta and the wild-type spike protein, SASA S vaccinated mice sera once again showed the highest levels of protection.
Slightly over half of the mice vaccinated only by the AdS+N vaccine did not show detectable levels of serum IgGs. However, an AdS+N booster vaccination following a SASA S vaccination does increase Cd4+ and CD8+ T cell response and mean antibody titers for this group were on par with SASA S homologous vaccination. This was most obvious with CD8+ T cells but could be seen in both. These responses were similar once the Delta spike protein peptides were taken into account, but heterogenous vaccination showed an improvement in comparison to homogenous. Once again, only mice that had been at least part vaccinated by the AdS + N shot showed any antigen response to the nucleotide protein antigen, and responses were similar whether this was homologous or heterogeneous vaccination.
The ELISpot tests showed that animals receiving heterologous vaccination showed much higher levels of spike protein-reactive IFN-gamma-secreting T cells than all groups apart from the SASA S homologous – which was only slightly lower. Against the nucleotide protein, the IFN-gamma response was similar for both homologous vaccination with AdS + N and heterologous vaccination with both shots. The pseudovirus neutralization assays showed that homologous vaccination from either group could successfully neutralize all variants of pseudoviruses, but the AdS + N group showed a lower capacity for all strains. The heterologous vaccination using AdS + N as a booster for SASA S once again proved more effective than using SASA S as a booster for AdS + N.
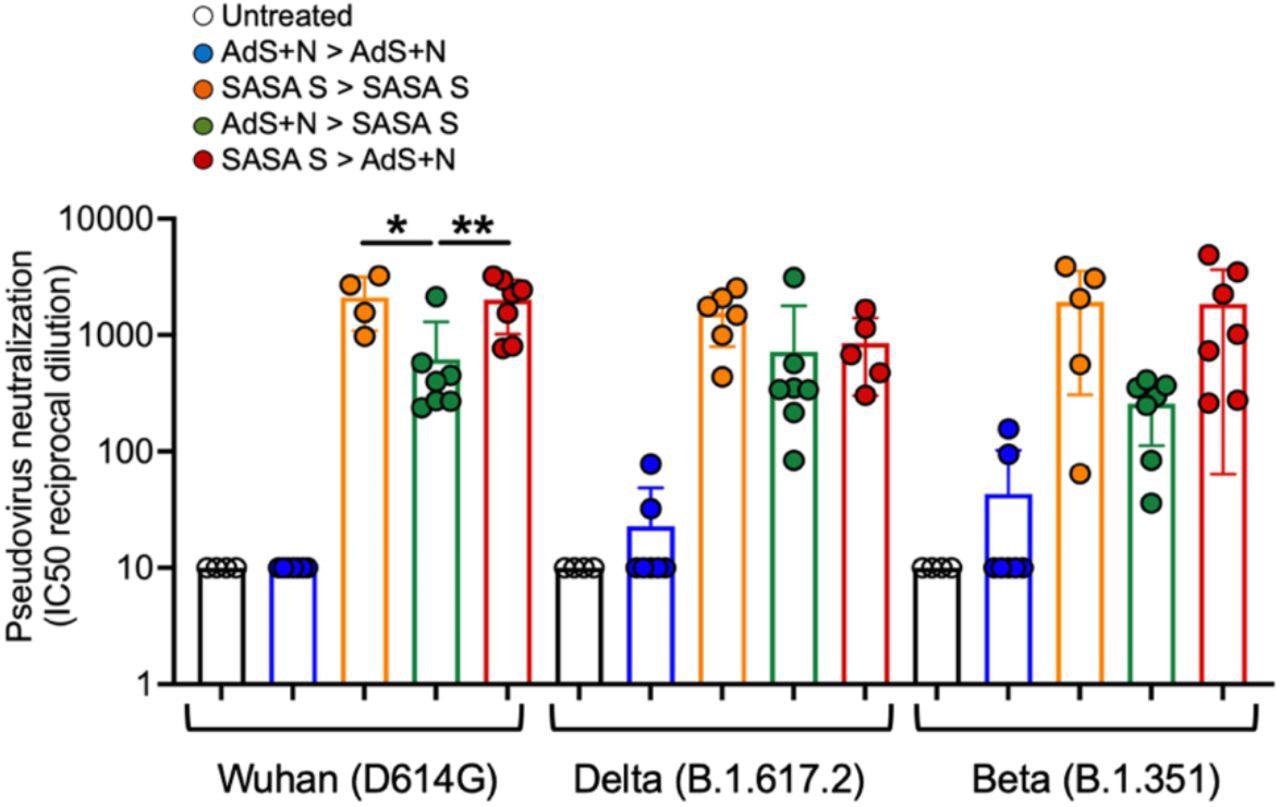 Sera from SASA S > AdS+N heterologously vaccinated mice neutralizes Wuhan, Delta, and Beta strain SARS-CoV-2 pseudovirus. Neutralization of Wuhan strain pseudovirus by sera from SASA S homologous and SASA S > AdS+N mice was significantly greater than that for AdS+N > SASA S mice (as well as untreated and AdS+N homologous). There were no significant differences between the SASA S homologous and the two heterologous groups for the Delta or Beta strain pseudoviruses. Statistical comparison of IC50 values for untreated and AdS+N homologous group mice with all/many values < the LOD was not performed. Statistical analyses were performed using one-way ANOVA and Tukey’s post-hoc comparison of SASA S homologous, AdS+N > SASA S, and SASA S > AdS+N groups to each other for each strain where *p ≤ 0.05 and **p < 0.01; and for the same group for each different strain. Data graphed as the mean and SEM.
Sera from SASA S > AdS+N heterologously vaccinated mice neutralizes Wuhan, Delta, and Beta strain SARS-CoV-2 pseudovirus. Neutralization of Wuhan strain pseudovirus by sera from SASA S homologous and SASA S > AdS+N mice was significantly greater than that for AdS+N > SASA S mice (as well as untreated and AdS+N homologous). There were no significant differences between the SASA S homologous and the two heterologous groups for the Delta or Beta strain pseudoviruses. Statistical comparison of IC50 values for untreated and AdS+N homologous group mice with all/many values < the LOD was not performed. Statistical analyses were performed using one-way ANOVA and Tukey’s post-hoc comparison of SASA S homologous, AdS+N > SASA S, and SASA S > AdS+N groups to each other for each strain where *p ≤ 0.05 and **p < 0.01; and for the same group for each different strain. Data graphed as the mean and SEM.
Conclusion
The authors highlight that their study supports the hypothesis that heterologous vaccination can provide an increased immune response against virus infection, supporting several other studies that have indicated similar events. While the heterologous vaccinations were not always significantly better than the homologous ones, they rarely induced a lower response and in some cases, such as in the CD4+ and CD8+ responses, showed significant improvements. This research could be invaluable for vaccine manufacturers and public health policymakers and could offer another avenue to help protect people from COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Adrian Rice, et al. (2021). Heterologous Vaccination with SARS-CoV-2 Spike saRNA Prime followed by DNA Dual-Antigen Boost Induces Robust Antibody and T-Cell Immunogenicity against both Wild Type and Delta Spike as well as Nucleocapsid Antigens. bioRxiv. doi: https://doi.org/10.1101/2021.11.29.470440 https://www.biorxiv.org/content/10.1101/2021.11.29.470440v1
- Peer reviewed and published scientific report.
Rice, Adrian, Mohit Verma, Emily Voigt, Peter Battisti, Sam Beaver, Sierra Reed, Kyle Dinkins, et al. 2022. “Heterologous SaRNA Prime, DNA Dual-Antigen Boost SARS-CoV-2 Vaccination Elicits Robust Cellular Immunogenicity and Cross-Variant Neutralizing Antibodies.” Frontiers in Immunology 13 (July). https://doi.org/10.3389/fimmu.2022.910136. https://www.frontiersin.org/articles/10.3389/fimmu.2022.910136.