Epigenetic mechanisms, such as DNA methylation, are crucial for both host immune responses to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and for pathogenesis and severity. Past studies have shown that SARS-CoV-2 infection alters the transcriptional landscape in severe coronavirus disease 2019 (COVID-19). However, insights into host DNA methylation states and longitudinal assessments of epigenetic clocks prior to and following COVID-19 are limited. Epigenetic clocks are distinct DNA methylation patterns used to derive epigenetic measures of biological aging.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In the absence of longitudinal epigenetic studies on COVID-19, it remains unclear whether all the aforementioned alterations in DNA methylation also occur in healthy individuals that recover from non-hospitalized COVID-19. Furthermore, it also remains unknown how these epigenetic mechanisms are impacted in individuals after they have received mRNA vaccination against COVID-19.
Study design
In a recent work posted to the medRxiv* pre-print server, a team of researchers performed longitudinal epigenetic studies on blood samples from individuals who had recovered from COVID-19. They investigated the alterations in the DNA methylation states, immune cell-type composition, and epigenetic clocks.
They examined longitudinal DNA methylation changes in the blood of 21 healthy participants before and following test-confirmed mild/moderate COVID-19 at a median timeframe of 8.35 weeks. Next, they analyzed longitudinal DNA methylation states, blood immune cell-type composition, and epigenetic clocks of 36 healthy participants before and after receiving two doses of mRNA-based COVID-19 vaccination.
Key findings
Aberrant DNA methylation related to immune dysfunction is often due to SARS-CoV-2 infection, and several past studies have reported such distinct blood-based DNA methylation as a potential biomarker of COVID-19. On observing blood-based DNA methylation changes associated with COVID-19 exposure in healthy participants, 261 differentially methylated, cytosine-guanine nucleotides linked by phosphate (CpGs), were identified at a false discovery rate (FDR) adjusted P-value < 0.05. The FDR is a statistical approach typically used in high-throughput experiments to correct random events that falsely appear significant. Hypermethylation at this locis was related to the caspase recruitment domain family member 14 (CARD14) gene, which encodes a protein interacting with B-cell lymphoma 10 (BCL10). BCL10 functions as a positive regulator of cell apoptosis.
The percentage change in DNA methylation following COVID-19 exposure indicates that a subset of DNA methylation changes related to COVID-19 exposure occurred due to cell-type compositional changes.
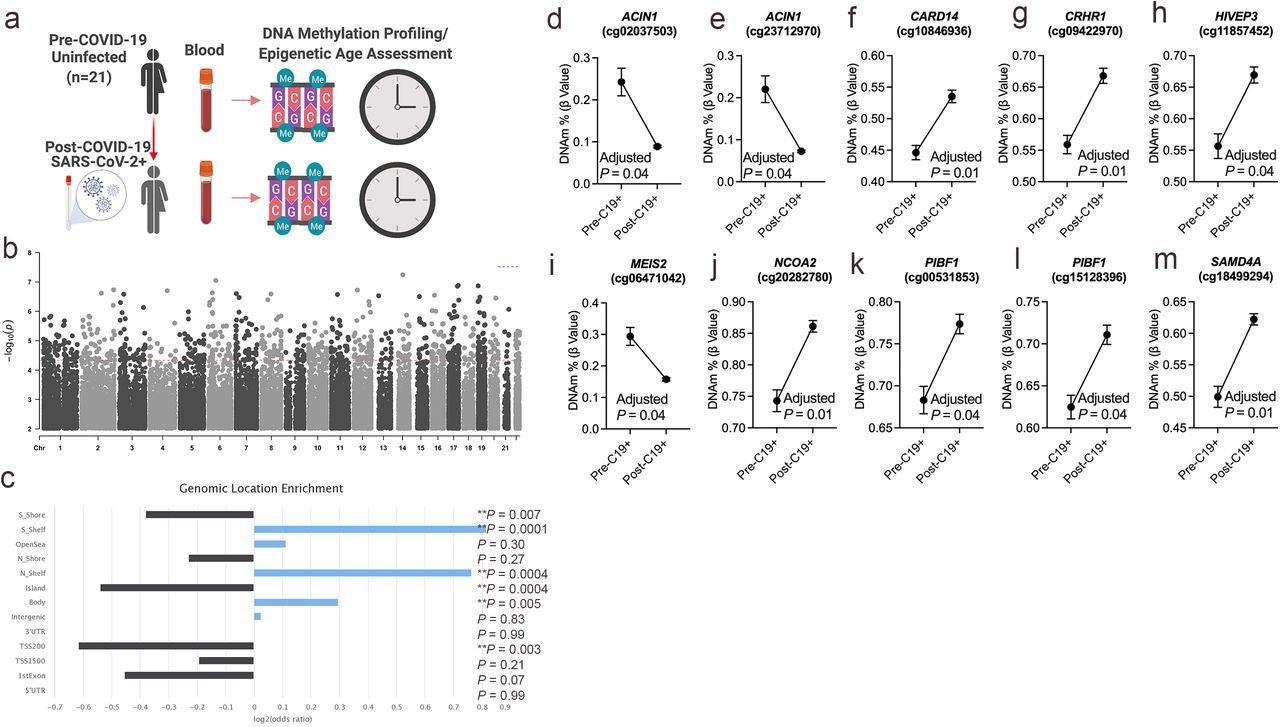 DNA methylation changes in blood associated with mild/moderate COVID-19. a. Study design of longitudinal assessment of DNA methylation profiles in 21 participants pre- and post-SARS-CoV-2 infection. b. Manhattan plot of differentially methylated loci (DML) associated with mild/moderate COVID-19. c. Bar graph of genomic enrichment of COVID-19 DML in 13 different categorized regions of the genome relative to gene and CpG island. The hypergeometric test was utilized to calculate P-value and odd ratio. d-m. Plots of COVID-19 DML displaying mean DNA methylation levels +/- SEM for CpGs associated with a gene ID. Adjusted P-value calculated utilizing Benjamini-Hochberg correction.
DNA methylation changes in blood associated with mild/moderate COVID-19. a. Study design of longitudinal assessment of DNA methylation profiles in 21 participants pre- and post-SARS-CoV-2 infection. b. Manhattan plot of differentially methylated loci (DML) associated with mild/moderate COVID-19. c. Bar graph of genomic enrichment of COVID-19 DML in 13 different categorized regions of the genome relative to gene and CpG island. The hypergeometric test was utilized to calculate P-value and odd ratio. d-m. Plots of COVID-19 DML displaying mean DNA methylation levels +/- SEM for CpGs associated with a gene ID. Adjusted P-value calculated utilizing Benjamini-Hochberg correction.
Notably, the differentially methylated CpGs associated with COVID-19 were enriched in transcriptional gene sets identified from published SARS-CoV-2 human, animal model, and in vitro infection studies. These findings suggested that these DNA methylation changes associated with COVID-19 also regulate and modulate the host's gene expression from infection. Together, these findings support the notion that distinct host DNA methylation states in circulating immune cells serve as a COVID-19 specific epigenetic signature. However, the durability of this COVID-19 epigenetic signature remains unclear and needs to be investigated in future studies.
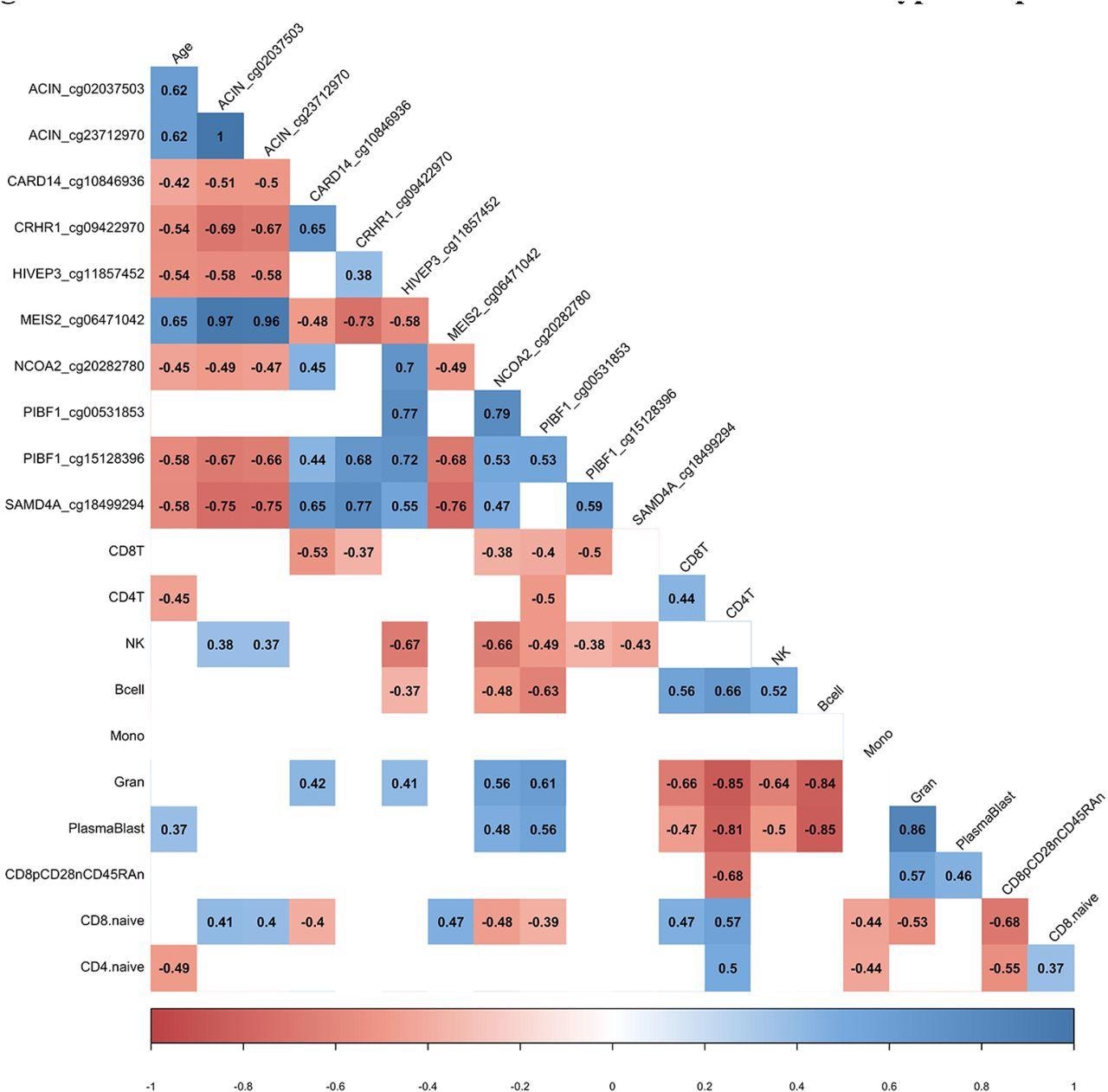 DML associated with COVID-19 relate to immune cell type composition. Correlogram plot of biological age, the change in DNA methylation levels for COVID-19 related DML, and the change in inferred immune cell type following COVID-19. Significant correlations displayed as solid box and Spearman’s rank correlation coefficient displayed.
DML associated with COVID-19 relate to immune cell type composition. Correlogram plot of biological age, the change in DNA methylation levels for COVID-19 related DML, and the change in inferred immune cell type following COVID-19. Significant correlations displayed as solid box and Spearman’s rank correlation coefficient displayed.
Unlike in the case of natural SARS-CoV-2 infection, epigenetic clocks significantly changed following Pfizer and Moderna mRNA vaccination against COVID-19 in individuals who were 50 years or older. The results revealed that principal component-based epigenetic clock estimates of PhenoAge and GrimAge significantly increased in people aged 50 years or older following infection by an average of 2.1 and 0.84 years. In contrast, PCPhenoAge significantly decreased in people 50 years or younger after infection by an average of 2.06 years. This observed divergence in individuals of different age bands suggests that epigenetic clock estimates pre- and post-COVID-19 vaccination was related to compositional changes in immune cells, such as B cells and plasmablasts, highlighting the potential utility of epigenetic clocks in capturing vaccine responses.
Conclusion
The study findings have important implications for research into the impact of COVID-19 and the mRNA vaccination on epigenetic aging in the immune system. Future research studies will examine whether COVID-19 and mRNA vaccine-related changes impacting the epigenetic age are biologically meaningful. In addition, they will have to establish the significance and durability of short-term changes in epigenetic aging. In the future, epigenetic clocks may serve as a potential biomarker of COVID-19 vaccine responses and track the need for booster shots due to waning COVID-19 immunity in older individuals.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources