After the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant in November 2021 in South Africa, the global medical community has raised concerns about the numerous mutations present in this variant of concern (VOC) and its ability to escape from vaccine-elicited immunity. Moreover, researchers anticipate that a large number of mutations in the receptor-binding domain (RBD) of the Omicron spike (S) protein would impact its transmissibility and affinity for the angiotensin-converting enzyme 2 (ACE2) receptor.
Study: SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. Image Credit: angellodeco / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the present study, researchers assessed the ability of plasma taken from vaccinated participants to neutralize the Omicron variant versus the D614G virus using a live virus neutralization assay. They also investigated whether the Omicron variant still needs to bind to the ACE2 receptor to infect host cells.
The researchers used a sequence-confirmed live Omicron virus isolate and a human lung cell line clone (H1299-ACE2) expressing the ACE2 receptor for testing virus neutralization. They also tested the growth of the viral isolate in parental H1299 cells that do not overexpress ACE2 and are not infectable with SARS-CoV-2.
In total, the plasma samples from 14 participants were included in the study. Hospitalized individuals with SARS-CoV-2 infection confirmed by polymerase chain reaction (PCR) and/or vaccinated individuals part of a prospective cohort study by the University of KwaZulu–Natal's Biomedical Research Ethics Committee donated blood samples for the analysis.
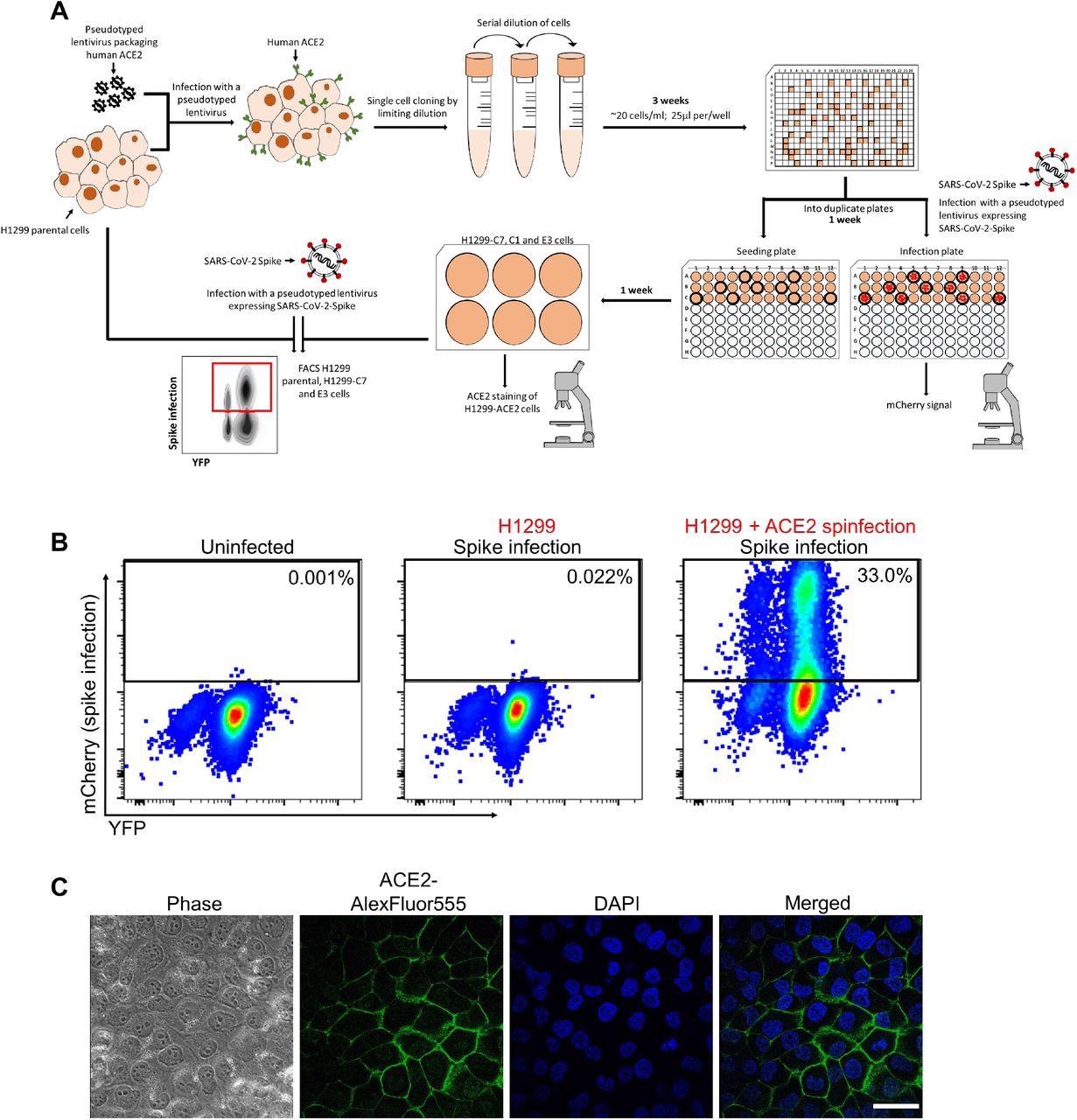 Generation of H1299-ACE2 clonal cell line. (A) The H1299 epithelial cell line with YFP labeled H2AZ was infected with the pHAGE2-EF1a-Int-ACE2 lentivector. Cells were single-cell cloned by limiting dilution in a 384 well plate. Clones were expanded into duplicate 96-well plates, where one plate was used to select infectable clones based on mCherry signal from infection with SARS-CoV-2 mCherry expressing spike pseudotyped lentivirus. Clones were chosen based on infectability and expanded from the non-infected replicate 96-well plate. (B) Flow cytometry plots of SARS-CoV-2 mCherry expressing spike pseudotyped lentivirus infection in H1299-ACE2 cells. (C) Images of H1299-ACE2 cells stained with anti-ACE2 antibody and DAPI. Note membrane localization of ACE2. Scale bar is 50μm.
Generation of H1299-ACE2 clonal cell line. (A) The H1299 epithelial cell line with YFP labeled H2AZ was infected with the pHAGE2-EF1a-Int-ACE2 lentivector. Cells were single-cell cloned by limiting dilution in a 384 well plate. Clones were expanded into duplicate 96-well plates, where one plate was used to select infectable clones based on mCherry signal from infection with SARS-CoV-2 mCherry expressing spike pseudotyped lentivirus. Clones were chosen based on infectability and expanded from the non-infected replicate 96-well plate. (B) Flow cytometry plots of SARS-CoV-2 mCherry expressing spike pseudotyped lentivirus infection in H1299-ACE2 cells. (C) Images of H1299-ACE2 cells stained with anti-ACE2 antibody and DAPI. Note membrane localization of ACE2. Scale bar is 50μm.
Of these 14 participants, six had no prior history of SARS-CoV-2 infection, as confirmed by the lack of nucleocapsid antibodies detected in their plasma samples. For two of these participants, researchers used samples from two-time points. The remaining six participants were infected with the ancestral D614G virus during the first SARS-CoV-2 infection wave in South Africa.
The researchers prepared the indexed paired-end libraries of genomic DNA and normalized, pooled, and denatured sequencing libraries before spiking with 1% PhiX. Next, the researchers aligned mapped reads to the reference sequences in the initial assembly from Genome Detective and subsequently filtered out low-quality mutations.
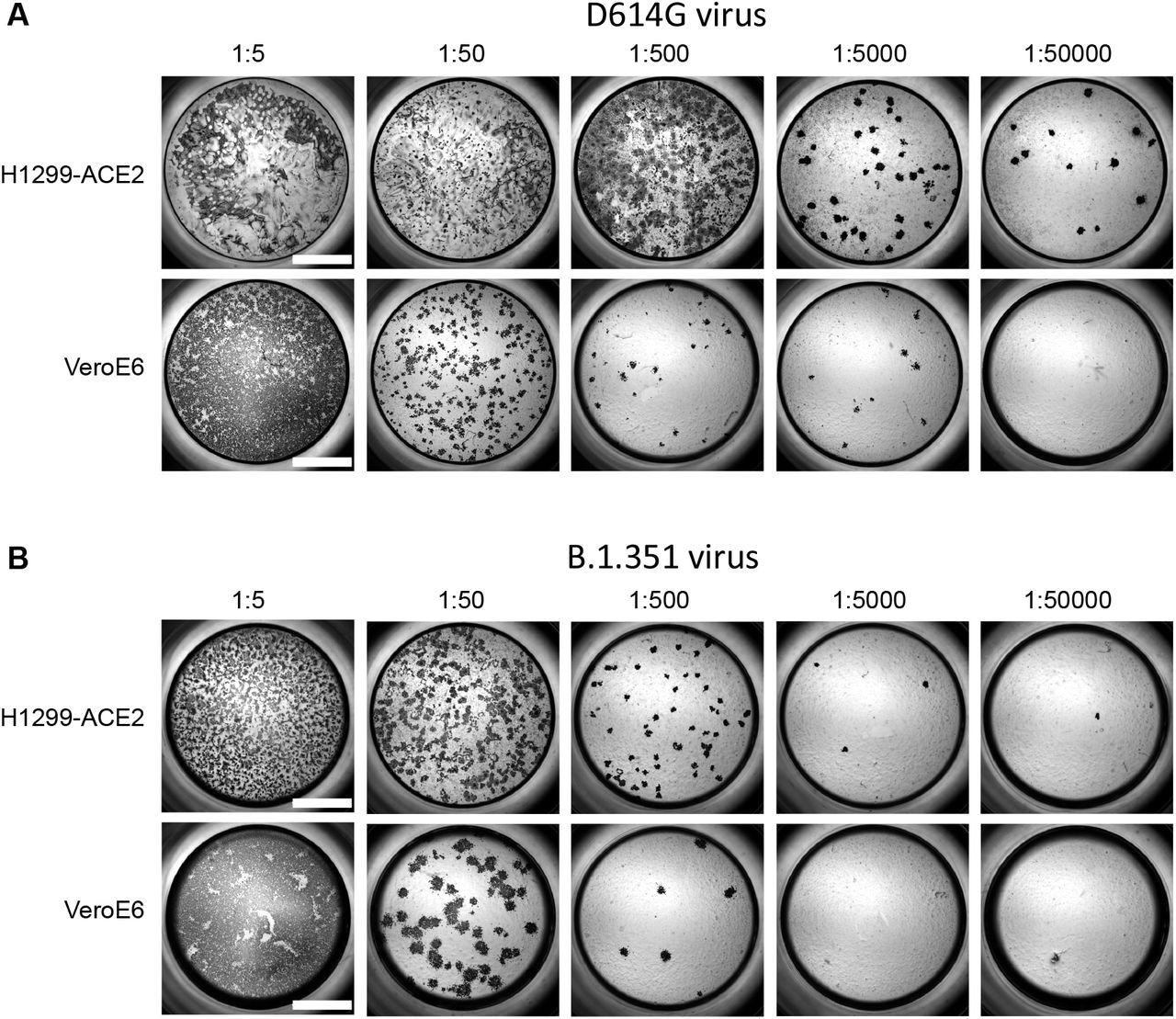 Comparison of SARS-CoV-2 infection in H1299-ACE2 and Vero-E6 cells. Both H1299-ACE2 and VeroE6 cells were infected with the same viral stock in the same experiment with D614G virus (A) or β virus (B) at different dilutions and a focus forming assay was performed. Scale bar is 2mm.
Comparison of SARS-CoV-2 infection in H1299-ACE2 and Vero-E6 cells. Both H1299-ACE2 and VeroE6 cells were infected with the same viral stock in the same experiment with D614G virus (A) or β virus (B) at different dilutions and a focus forming assay was performed. Scale bar is 2mm.
Neutralization data were estimated using the formula Tx=1/1+(D/ID50), where Tx is the number of foci normalized to the number of foci in the absence of plasma on the plate at dilution D. Comparatively, ID50 is the plasma dilution giving 50% neutralization. FRNT50 = 1/ID50 is the inverse of the plasma dilution required for a 50% reduction in infection foci number.
Study findings
The study results show that Omicron infected the ACE2-expressing cells but did not infect the parental H1299 cells, thus indicating that its ability to escape antibody neutralization elicited by the Pfizer BNT162b2 messenger ribonucleic acid (mRNA) vaccine depends on ACE2 concentration levels and ACE2 is necessary for Omicron entry and infection establishment.
For the D614G virus, the geometric mean titer (GMT) FRNT50 was 1,321 and these samples strongly neutralized the D614G virus. For the Omicron variant, GMT for the same samples was 32, thereby indicating a 41-fold decline in FRNT50.
However, five of the previously infected participants showed relatively high neutralization titers against Omicron, thus indicating that immune escape was incomplete. Comparatively, previous data confirmed that the Beta variant showed significant immune escape from BNT162b2 in a live virus neutralization assay, with a nearly 3-fold reduction in FRNT50.
Conclusions
The study results show that the new SARS-CoV-2 Omicron variant extensively escapes vaccine-elicited neutralization; however, the escape was incomplete in participants who had higher FRNT50 from a previous infection. Therefore, an earlier infection followed by vaccination or vaccine booster dose is expected to increase the neutralization level and confer protection against severe disease from SARS-CoV-2 Omicron infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Cele, S., Jackson, L., Khan, K., et al. (2021). SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. doi:10.1101/2021.12.08.21267417. https://www.medrxiv.org/content/10.1101/2021.12.08.21267417v2
- Peer reviewed and published scientific report.
Cele, Sandile, Laurelle Jackson, David S. Khoury, Khadija Khan, Thandeka Moyo-Gwete, Houriiyah Tegally, James Emmanuel San, et al. 2022. “Omicron Extensively but Incompletely Escapes Pfizer BNT162b2 Neutralization.” Nature 602 (7898): 1–3. https://doi.org/10.1038/s41586-021-04387-1. https://www.nature.com/articles/s41586-021-04387-1.