Evaluating the characteristics of emerging SARS-CoV-2 variants of concern (VOCs) plays an important role in pandemic risk assessment. The faster growth of variants may be either due to a large number of secondary infections, which is otherwise known as a transmissibility advantage, or better timing of secondary infections or generation time. Previous studies have primarily focused on deriving the transmissibility advantage considering the generation time remained a constant.
About the study
The authors of the present study developed a mathematical model to investigate the variation of the growth advantage in the epidemiological context during different generation times of SARS-CoV-2 variants. The relationship between infectivity profile and growth rate was used to conceptually explore the selection of infectivity profiles.
The rate of infectious contact in the community is known as transmission and depends upon interventions and factors such as social distancing, mask-wearing, population immunity, and seasonal effects. The time-varying reproduction number of the historical strain was considered as a proxy for transmission.
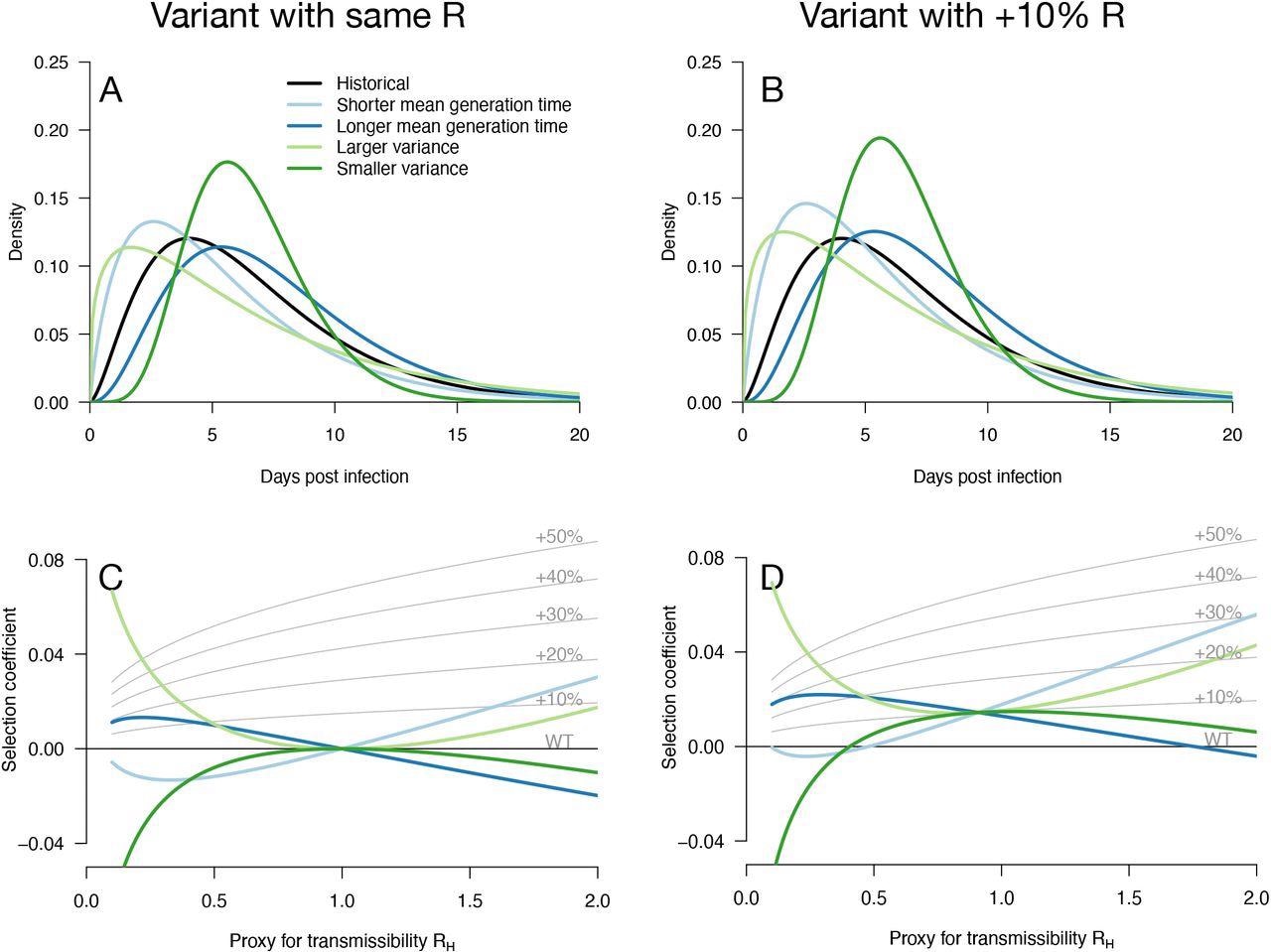 Variation of the selection coefficient as a function of transmission, for several infectivity profiles of emerging variants. Panels A, B show several variant infectivity profiles with the same effective reproduction number as historical strains (A) or an effective reproduction number increased by +10% (B). For variants with shorter or longer mean generation time, the relative mean generation time is -15% or +15% compared to historical strains. For variants with shorter or longer sd in generation time, the relative sd in generation time is -40% or +40% compared to historical strains. The Panels C, D show for these variants, the selection coefficient as a function of the proxy for transmission (effective reproduction number of the historical strains). The gray lines show the selection coefficient if the variant had the same infectivity profile as historical strains, only increased uniformly by a constant as commonly assumed.
Variation of the selection coefficient as a function of transmission, for several infectivity profiles of emerging variants. Panels A, B show several variant infectivity profiles with the same effective reproduction number as historical strains (A) or an effective reproduction number increased by +10% (B). For variants with shorter or longer mean generation time, the relative mean generation time is -15% or +15% compared to historical strains. For variants with shorter or longer sd in generation time, the relative sd in generation time is -40% or +40% compared to historical strains. The Panels C, D show for these variants, the selection coefficient as a function of the proxy for transmission (effective reproduction number of the historical strains). The gray lines show the selection coefficient if the variant had the same infectivity profile as historical strains, only increased uniformly by a constant as commonly assumed.
The conceptual exploration suggested that changes in the infectivity profile associated with an emerging variant can be inferred along with its growth rate when transmission changes over time. This approach was applied to the frequency of the SARS-CoV-2 Alpha and Delta variants over time in England during the Fall of 2020 and Spring of 2021.
The authors considered a situation in which historical strains H, which included a variant or a set of strains with similar properties, were circulating for a short span of time and were challenged by an emerging variant E. During the investigation of the rise of variant E relative to historical strains H, the exponential growth rates of H and E strains were observed and translated into a growth advantage or disadvantage.
 Epidemiological and evolutionary trajectories of several types of emerging variants competing with historical strains. Panels A, B show epidemiological dynamics (daily cases number of historical strains in black, of variants in colours). Historical strains display several slightly different curves when competing with each of the variants because of the weak competition brought about by the build-up of population immunity. Panels C, D, show evolutionary trajectories, with daily variant frequency as coloured lines. The light gray curves show the frequency dynamics of variants with a +10% to +50% transmissibility advantage, with the same generation time as historical strains. The inset shows the transmission through time in these simulations, given by the basic reproduction number of historical strains.
Epidemiological and evolutionary trajectories of several types of emerging variants competing with historical strains. Panels A, B show epidemiological dynamics (daily cases number of historical strains in black, of variants in colours). Historical strains display several slightly different curves when competing with each of the variants because of the weak competition brought about by the build-up of population immunity. Panels C, D, show evolutionary trajectories, with daily variant frequency as coloured lines. The light gray curves show the frequency dynamics of variants with a +10% to +50% transmissibility advantage, with the same generation time as historical strains. The inset shows the transmission through time in these simulations, given by the basic reproduction number of historical strains.
When the historical strains grew with exponential rate rH and the emerging variant grew with rate rE, the logit of the frequency of the emerging variant grew linearly with slope, which is known as the growth advantage or selection coefficient.
The assumption was that both variant and historical strains shared the same distribution of generation time. Therefore, the transmissibility advantage, which is the effective reproduction number of the emerging variant relative to historical strains, could be inferred.
Study findings
The key finding of this study was that the transmission levels reflected in the reproduction number change selection on different variants. In terms of high transmission and growth rate, most infections are recent and early transmission is more advantageous.
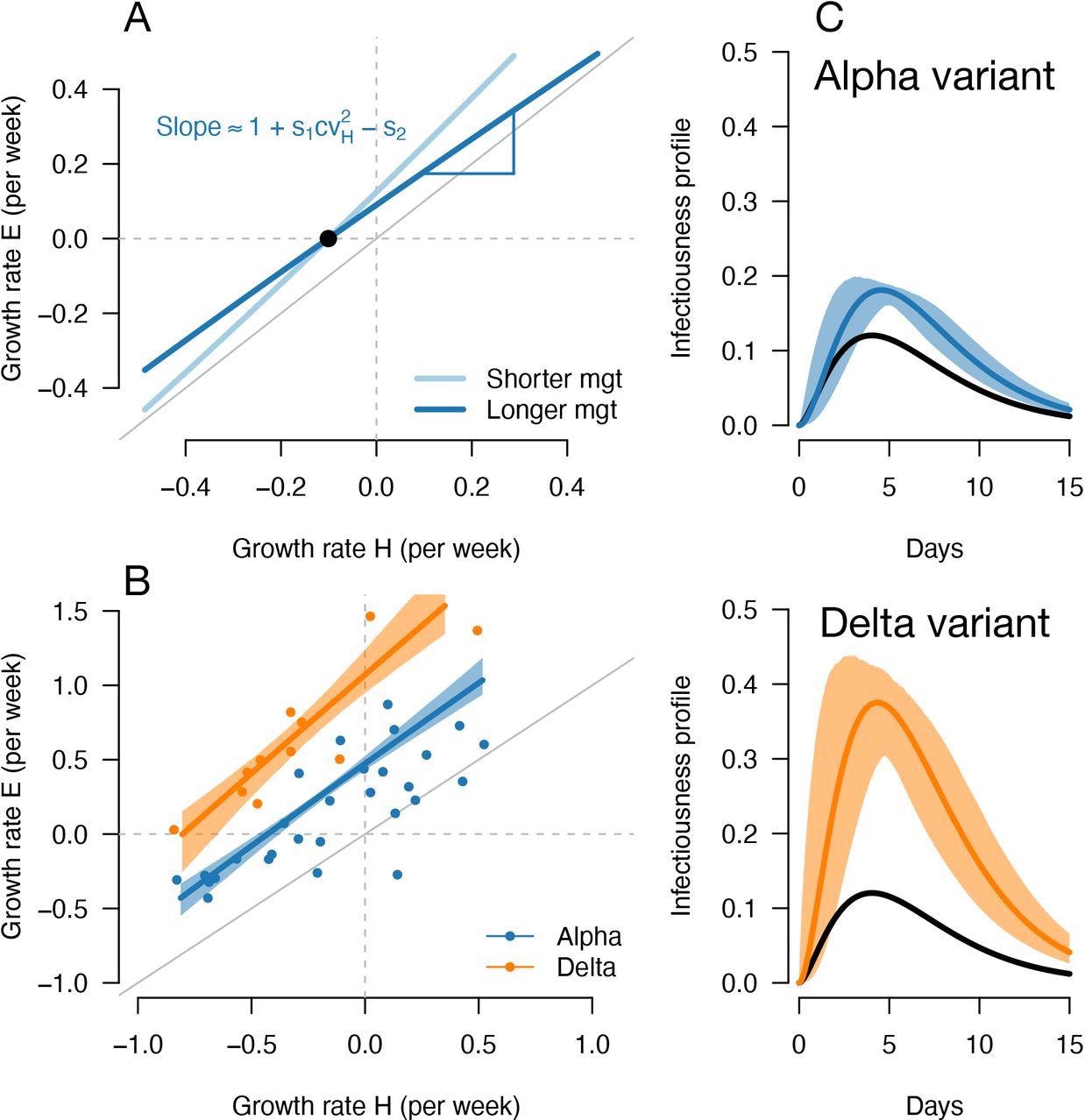 A, Geometric intuition for how the (rH, rE) relationship depends on the mean generation time for hypothetical variants with transmissibility advantage s1=0.1 and relative mean generation time of s2= 0.15 and s2=+0.15 (as in Fig. 1, 2). B, the correlation between estimated growth rates of historical strains and variant in England. For Alpha (dataset 1) this is a temporal correlation in England over weeks starting 08/09/2020 to 16/03/2021. For Delta (dataset 3) this is a spatial correlation across European countries. The curves with confidence intervals are the model fits for each variant and show that the selection coefficient (the vertical distance to the bisector) is roughly constant across epidemiological conditions, indicative that the mean generation time is not altered in these variants. C, the inferred infectivity profile of each variant with confidence intervals, together with the infectivity profile of historical strains circulating before the rise of the Alpha variant (black curve).
A, Geometric intuition for how the (rH, rE) relationship depends on the mean generation time for hypothetical variants with transmissibility advantage s1=0.1 and relative mean generation time of s2= 0.15 and s2=+0.15 (as in Fig. 1, 2). B, the correlation between estimated growth rates of historical strains and variant in England. For Alpha (dataset 1) this is a temporal correlation in England over weeks starting 08/09/2020 to 16/03/2021. For Delta (dataset 3) this is a spatial correlation across European countries. The curves with confidence intervals are the model fits for each variant and show that the selection coefficient (the vertical distance to the bisector) is roughly constant across epidemiological conditions, indicative that the mean generation time is not altered in these variants. C, the inferred infectivity profile of each variant with confidence intervals, together with the infectivity profile of historical strains circulating before the rise of the Alpha variant (black curve).
However, during a slow epidemic with low transmission rates, infections are older and late transmission is more advantageous. The selection coefficient may increase or decrease with the transmission or can even be a non-monotonous function of transmission.
Another important finding was about the understanding of the infectivity profile with the variation of the selection coefficient with the transmission. The study revealed that the SARS-CoV-2 Alpha variant had a transmissibility advantage of +54% relative to historical strains, whereas the Delta variant had a +140% additional advantage compared to the Alpha variants.
The generation time of both variants was similar to that of historical strains. The study data may help interpret the variant frequency dynamics, which are determined by the selection coefficient.
Limitations and conclusions
The current study did not consider the influence of interventions that can shorten the distribution of generation time such as isolation of positive cases and contact tracing, which also potentially change over time. The study also did not consider the impact of other changes in the environment, such as selection on vaccine escape variants, which may increase over time in the context of rapid vaccination of the population.
In conclusion, the researchers developed an analytical framework to select variants that can modify the transmission of an infectious pathogen. The findings from this study will help understand variant dynamics at a time when several SARS-CoV-2 VOCs are in circulation and new ones are emerging.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.