The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant was initially discovered in South Africa and has since spread throughout the world. The widespread transmission of the Omicron variant has been linked to a rapid increase in the frequency of cases, with current research demonstrating that neutralizing antibody responses are significantly evaded against this new strain of SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Current knowledge on Omicron
In the United Kingdom, Omicron appears to be competing with the previously dominant Delta variant. This could be due to an advantage in both the vaccinated and previously exposed populations and/or greater replication. However, there is a scarcity of information available on the replication characteristics of this variant.
The SARS-CoV-2 Delta spike protein has more efficient cell-cell fusion kinetics as compared to the wild-type Wuhan strain. Furthermore, the pathogenicity of this strain has been linked to alterations in the polybasic cleavage site (PBCS). The Omicron variant is projected to be extremely infectious and fit because of three mutations in the furin cleavage site region including P681H, H655Y, and N679K.
In a recent study published on the preprint server bioRxiv*, the Omicron spike is found to be relatively poorly cleaved and impaired in mediating cell-cell fusion and syncytia formation, contrary to predictions based on mutational profiling of the PBCS region. This decreased cleavage is also linked to poorer entry into target lung organoids or cell lines expressing endogenous receptor levels.
Furthermore, the authors show that, as predicted by mutational profiling, the SARS-CoV-2 Omicron variant has significantly lower sensitivity to neutralizing antibodies and that the vaccine sera from individuals who have received the AstraZeneca (AZ) vaccine have lower titers as compared to those who have received either of the messenger ribonucleic acid (mRNA) vaccines. However, robust titers against Omicron can be achieved soon after the third dose of an mRNA vaccine, thus supporting third dose 'boosting' strategies.
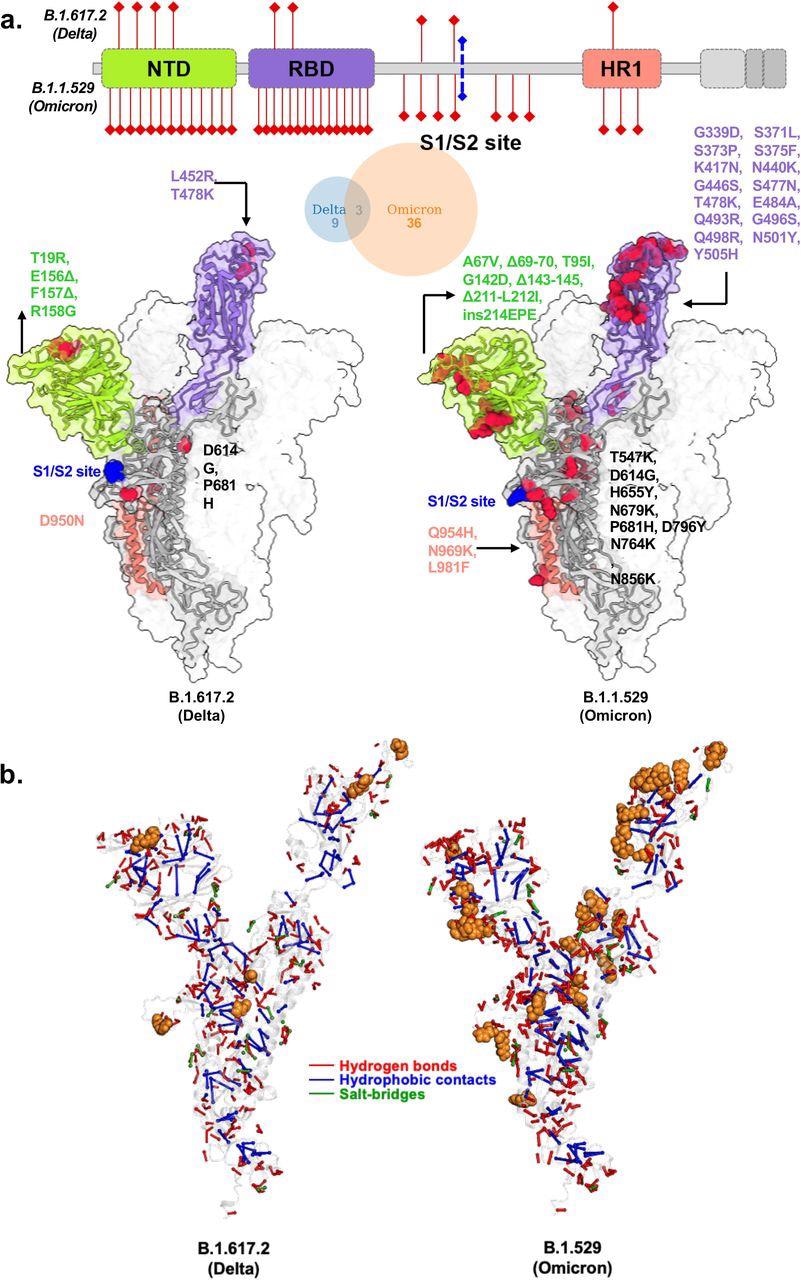
Structural models of SARS-CoV-2 Delta and Omicron spike variants and their corresponding protein intramolecular network. a. Variant structures for Delta and omicron of spike protein displaying mutational sites in red color. The schematic highlights differences in domain-wise mutations across protein length. b. The non-covalent interactions, including hydrogen bonds, hydrophobic contacts, and salt bridges are shown for Delta and omicron variants in red, blue, and green, respectively. The interactions were calculated on 100 ns molecular dynamics simulation and interactions with >50% persistence are shown. For clarity only unique hydrogen bonds specific to each variant spike protein are marked in red for one chain. The mutations present in Delta and Omicron are marked in.
About the study
In the current study, the researchers created pseudovirus (PV) particles by synthesizing codon-optimized spike expression plasmids for Omicron and Delta spike proteins. Longitudinal serum samples were collected from people who had received either the Pfizer-BioNTech BNT162b2 or AZ ChAdOx-1 vaccines.
When compared to the Delta variant, the authors observed a ten-fold loss of neutralization against the Omicron variant after the second dose. Indeed, the majority of people who received two doses of ChAdOx-1 had no detectable Omicron neutralization.
The authors also noticed a waning over time since vaccinees had received their second dose. Both groups were given a third dose of BNT162b2, allowing comparisons to be made to the response to this boosting dose. All tested SARS-CoV-2 variants showed similar magnitude increases in neutralization, thus indicating an increased breadth of responses as well as titer levels.
Mutations in the PBCS at P681 have been found in several SARS-CoV-2 lineages, most notably the Alpha and Delta variants. The authors previously demonstrated that these P681 mutant spikes had significantly higher fusogenic potential than a D614G wild-type spike. Notably, the Omicron variant has the P681H mutation, as well as the 679 and 655 mutations.
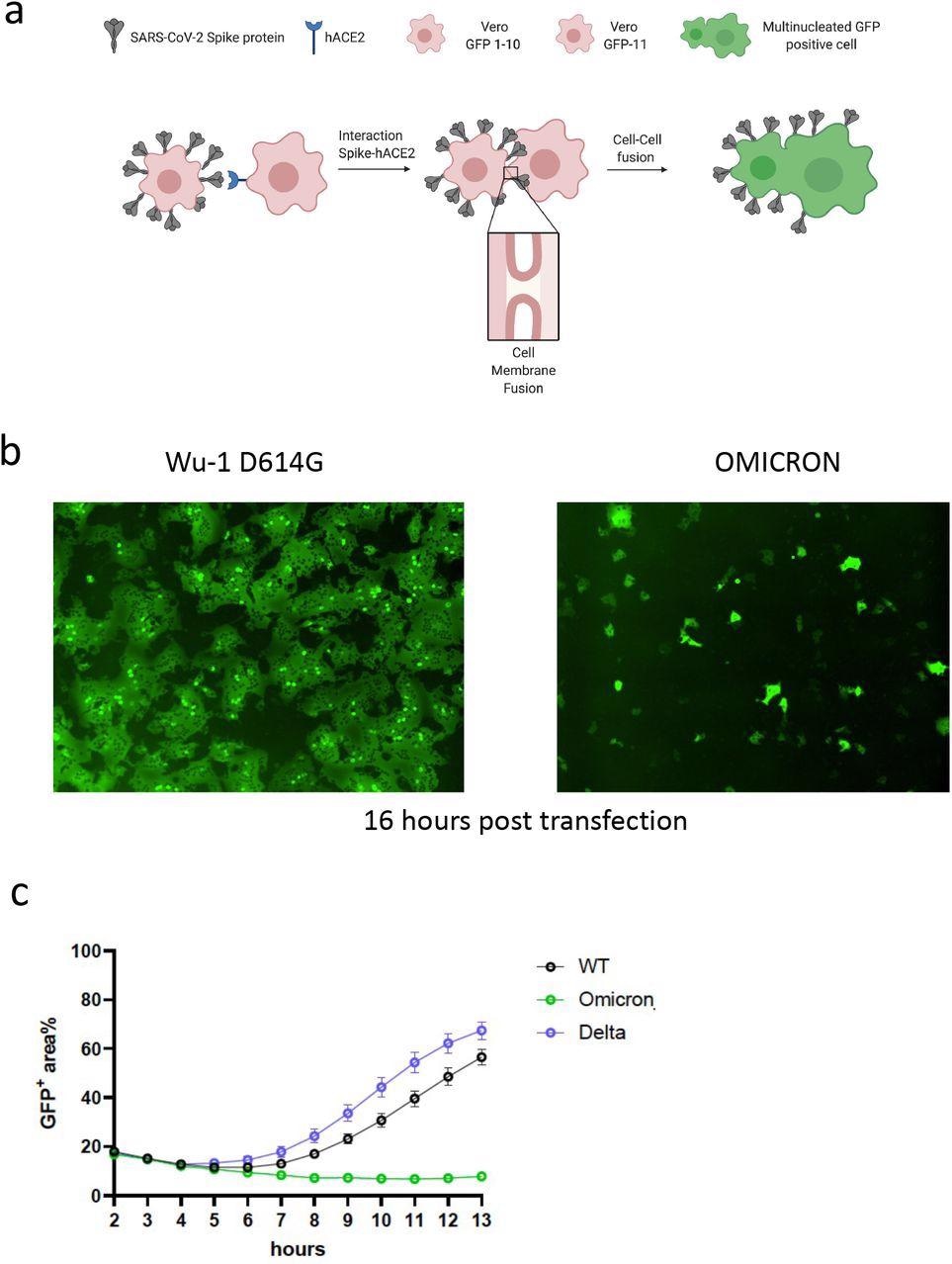
SARS-CoV-2 Omicron variant spike confers impaired cell-cell fusion activity. a. Schematic of cell-cell fusion assay. b. Reconstructed images at 16 hours of GFP+ syncytia. c. Quantification of cell-cell fusion kinetics showing percentage of green area to total cell area over time (WT is Wuhan-1 D614G). Mean is plotted with error bars representing SEM. Data are representative of at least two independent experiments.
The authors used a split green fluorescent protein (GFP) system to monitor cell-cell fusion in real-time while testing the Omicron spike protein. The authors transfected spike-bearing plasmids into 293T cells expressing GFP1-10 and mixed them with Vero cells stably expressing GFP-11 to allow the GFP signal to be detected and measured upon cell-cell fusion. The authors observed increased fusion for the Delta variant when compared to the previous D614G wild-type spike. Despite being expressed, the Omicron spike resulted in very poor fusion.
The PV system was also used to infect primary three-dimensional (3D) lung alveolar organoids and Calu-3 lung cells expressing endogenous levels of angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), as well as the A549 lung cell line transduced with ACE2 and TMPRSS2. When PV virions were probed for the spike protein, the Omicron spike protein was predominantly in the uncleaved form, which was in contrast to the Delta and wild-type spike protein appearances.
The authors discovered that the Omicron variant has a variable entry efficiency when compared to Delta and wild-type D614G. The Omicron variant inhibited SARS-CoV-2 infection in organoids and Calu-3 lung cells as compared to Delta and wild-type D614G. Calu-3, surprisingly, does not express Cathepsin L, thereby implying that entry is entirely dependent on the plasma membrane.
Implications
The findings from the current study indicate that the SARS-CoV-2 Omicron variant has acquired immune evasion properties while impairing syncytia formation and cell entry in lung cells, which may have pathogenic implications. Critically, ChAdOx-1 is widely used in low-income settings where third doses with mRNA are in short supply; therefore, Omicron may contribute to severe disease in such settings.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Meng, B., Ferreira, I., Abdullahi, A., et al. (2021). SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion. bioRxiv. doi:10.1101/2021.12.17.473248. https://www.biorxiv.org/content/10.1101/2021.12.17.473248v2.
- Peer reviewed and published scientific report.
Meng, Bo, Adam Abdullahi, Isabella A. T. M. Ferreira, Niluka Goonawardane, Akatsuki Saito, Izumi Kimura, Daichi Yamasoba, et al. 2022. “Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Tropism and Fusogenicity.” Nature, February. https://doi.org/10.1038/s41586-022-04474-x. https://www.nature.com/articles/s41586-022-04474-x.