Despite the availability of effective vaccines, several SARS-CoV-2 variants of concern (VOCs) are circulating globally with mutations in the spike (S) protein, some of which are associated with increased transmissibility and immune escape. As a result, the Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529) VOCs, which are more resistant to vaccine-elicited immunity, have become predominant over the wildtype strain and the previously dominant Alpha (B.1.1.7 71) variant.

Study: Molecular probes of spike ectodomain and its subdomains for SARS-CoV-2 variants, Alpha through Omicron. Image Credit: Design_Cells / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the current study, the researchers designed biotin-labeled molecular probes of wildtype SARS-CoV-2 S ectodomain and subdomains for identifying potent neutralizing antibodies (NAbs) targeting S specific regions of several VOCs, such as Alpha, Beta, Gamma, Epsilon, Iota, Kappa, Delta, Lambda, Mu, and Omicron variants.
The researchers synthesized 14 N-terminal domain (NTD) probes, 12 receptor-binding domain (RBD) probes, and 12 RBD with S subdomain 1 (RBD-SD1) probes, alongside their WA-1 strain counterpart, similar in expression to prefusion-stabilized spike ectodomain (S2P) probe. They also estimated the overall yield, physical properties, and antigenicity of these probes to quantify immune responses against S.
The researchers evaluated the biotinylation and binding affinity of these probes to the antibody panel using bio-layer interferometry (BLI) and assessed the impact of incorporated VOC mutations on vaccine effectiveness for finding correlates of elicited immune responses with neutralization. Next, the researchers functionally validated these molecular probes by using yeast expressing a panel of nine SARS-CoV-2 S-binding antibodies. The sorting capabilities of these antibodies were confirmed using yeast displaying libraries of convalescent plasma from individuals previously infected with SARS-CoV-2. Lastly, these probes were deposited to Addgene for dissemination.
Study findings
The molecular probe constructs incorporated an N terminal purification tag denoted as ‘scFc3C,’ a site-specific protease-cleavage site, an antigen sequence from SARS-CoV-2 variants having the entire S ectodomain (residues 14 to 1208) or S2P, NTD, RBD, and RBD-SD1, and 10lnQQ-AVI, which is a 10-residue linker, followed by a biotin ligase-specific sequence GLNDIFEAQKIEWHE at the C terminal.
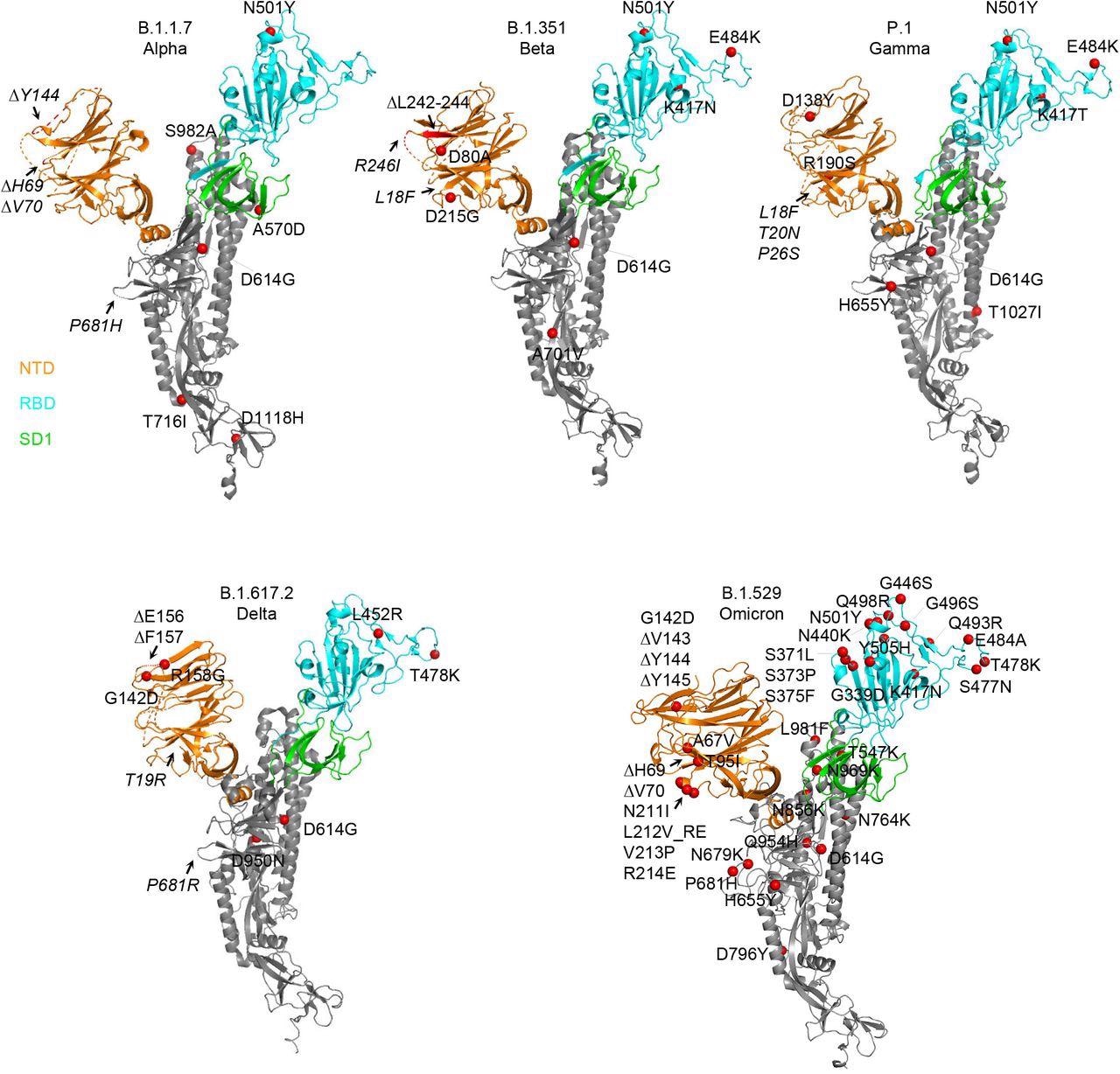
Structural modeling reveals a persistent D614G mutation, shared RBD mutations, and distinctive NTD mutations in five variants of concern. Structural models of a SARS-CoV-2 spike protomer from B.1.1.7, B.1.351, P.1, B.1.617.2, and B.1.1.529 variants. NTD, RBD, and SD1 domains are shown in orange, cyan, and green, respectively. Mutations are highlighted in the structural diagrams with a red sphere at Cα. Mutations at positions that are not resolved in the cryo-EM structure are labeled in italics with arrows pointing to the disordered regions (shown with dashed lines).
Several previous studies on SARS-CoV-2 antibodies have shown the critical role of the epitopes of RBD and NTD antibodies in immunogenicity, which is why they are widely used for the isolation and characterization of NAbs. While all purified NTD probes did not react to S2-directed or RBD-directed antibodies, they clearly showed high responses to antibody S652-118, thereby indicating that the epitope of S652-118 was conserved among SARS-CoV, WA-1 SARS-CoV-2, and all SARS-CoV-2 variants investigated in this study. In addition, the low reactivity of S652-118 to the S2P probes indicated its restricted access to the S652-118 epitope by other domains in the S2P trimer.
RBD-binding NAb CR3022 and the non-neutralizing antibody S652-109 showed lower responses to all the S2P variants, except for P.1 S2P. Other RBD-targeting antibodies, such as A23-58.1, A19-61.1, and B1-182.1, bound to all S2P probes, except to Omicron, whereas A19-46.1 recognized only S2P probes without L452 mutation.
The NTD-specific NAb S652-118 showed a similar pattern; however, the NTD-specific antibody S652-112, which recognized an epitope in S2, bound to the Delta S2P but not with the other variants with no mutations in their S2 subunit (B.1.429, and B.1.618). The three NTD-directed Nabs recognized most of the variants, except two Delta variants, B.1.617.2 and AY.1, and the Lambda variant C.37, thus indicating its epitope is unaffected by mutations in the NTD region of most of the variants.
As observed with S2P probes, all RBD and RBD-SD1 probes showed high responses to RBD-antibodies, except antibodies A19-46.1 and A19-61.1. A19-46.1, which could not bind probes with L452R/L452Q mutation. On the other hand, unlike the Omicron S2P, the Omicron RBD lost binding to only class 3 RBD antibody A19-61.1.
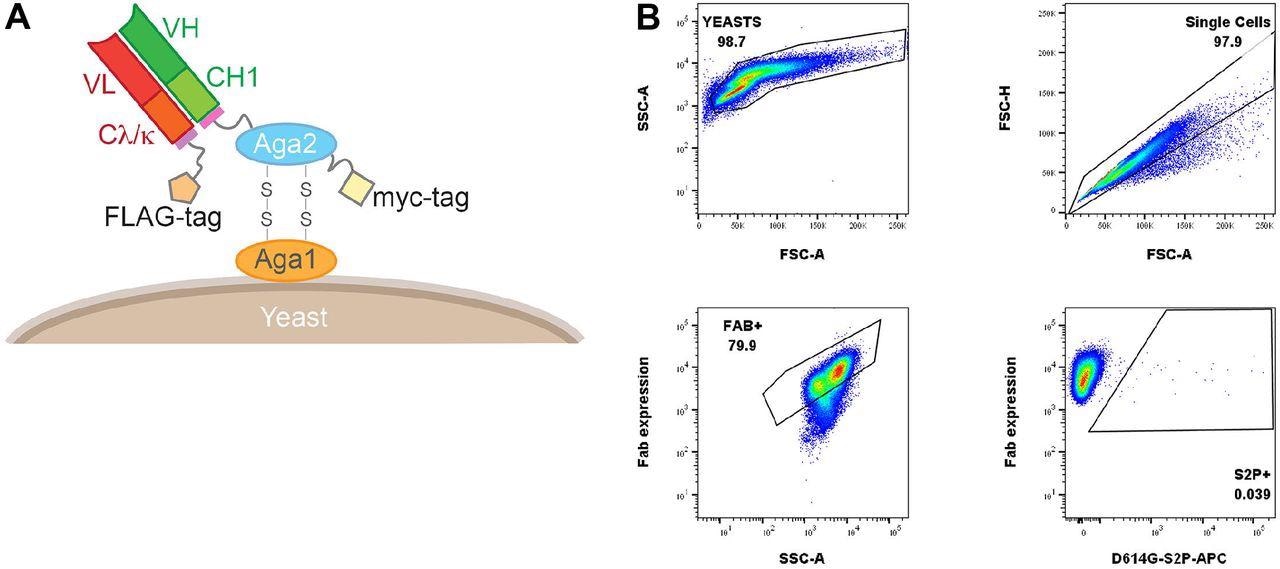
Yeast Fab display and gating tree for yeast display analysis of probe binding. (A) Saccharomyces cerevisiae strain AWY101 transfected with yeast display vector and Fab display is induced by incubating yeast in galactose containing media. The presence of Fab expressed on the yeast surface can be detected by staining with an anti-Flag antibody and analyzing using flow cytometry. (B) Induced yeast bearing Fabs of interested are analyzed by the indicated gating strategy. Singlets are analyzed for Fab expression and the proportion of probe binding determined within this population of yeast. Shown is a representative data.
Conclusions
Taken together, the study findings demonstrate the vital role biotin-labeled molecular probes for SARS-CoV-2 VOCs could play in antibody discovery and immune evaluation. In addition, these molecular probes could be helpful for diagnosing sera reactivity, as sensitive markers of SARS-CoV-2 infection, and in pathogenesis to delineate susceptible cells that SARS-CoV-2 might infect. However, when used for identifying the S regions recognized by antibodies, the study results showed that the probes defining RBD and RBD-SD1 were somewhat redundant due to considerable overlap between these two probe regions.
Furthermore, the study results showed the overall yields in the range of 1.5-5.9 mg/L, 3.7-11.3 mg/L, and 3.8-18.3 mg/L for NTD, RBD, and RBD-SD1 probes, respectively. The poor yield of Omicron RBD (0.18 mg/L) in comparison to the Omicron RBD-SD1 (0.88 mg/L) demonstrates the additional utility of the Omicron RBD-SD1 construct.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Teng, I. T., Nazzari, A. f., Chloe, M., et al. (2021). Molecular probes of spike ectodomain and its subdomains for SARS-CoV-2 variants, Alpha through Omicron. bioRxiv. doi:10.1101/2021.12.29.474491. https://www.biorxiv.org/content/10.1101/2021.12.29.474491v1
- Peer reviewed and published scientific report.
Teng, I-Ting, Alexandra F. Nazzari, Misook Choe, Tracy Liu, Matheus Oliveira de Souza, Yuliya Petrova, Yaroslav Tsybovsky, et al. 2022. “Molecular Probes of Spike Ectodomain and Its Subdomains for SARS-CoV-2 Variants, Alpha through Omicron.” Edited by Kevin A. Henry. PLOS ONE 17 (5): e0268767. https://doi.org/10.1371/journal.pone.0268767. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0268767.