
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
There is a critical need to discover effective antivirals against SARS-CoV-2 that could complement the current vaccination strategy to combat the ongoing pandemic caused by this virus. To address this need, the present study focused on a human lung cell line-based model that could reproduce the SARS-CoV-2 life cycle, be conducive for studying virus-host interactions and facilitate the discovery of novel antiviral drugs.
About the study
SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) in host cells as a receptor for entry and transmembrane protease serine 2 (TMPRSS2) to prime the spike (S) protein. The researchers of the present study transduced lentiviral ACE2 and TMPRSS2 genes sequentially into the A549 cells. Following puromycin selection, they tested over 50 clones for SARS-CoV-2 virus infection.
The parental clone 43.20 showed low levels of ACE2 expression but exhibited an infectivity rate of about 20%. As a result, the researchers selected this clone for further ACE2 receptor enrichment by cell sorting. To this end, the sorted cell population exhibiting a high level of ACE2 expression was referred to as ACE2plus, which was also permissible to SARS-CoV-2 replication.
The team performed a comparative infection with WA1/2020 isolate and recombinant virus (icSARS-CoV-2mNG) in ACE2plus and Vero E6 cells. Wild-type infected ACE2plus cells exhibited infection rates of about 60%, while Vero E6 cells showed infectivity rates of about 70%.
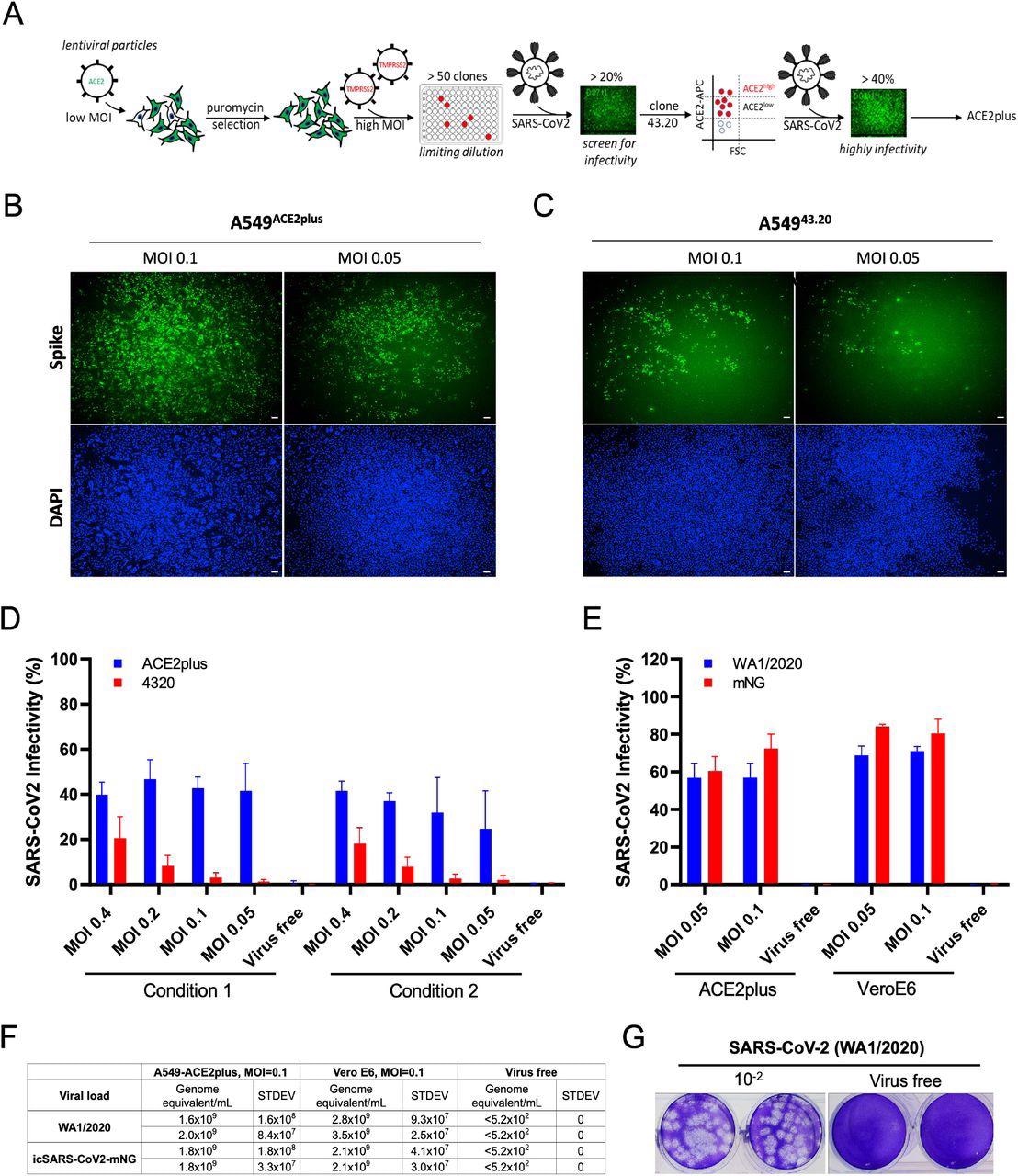
Establishment of highly permissive ACE2plus cell model for SARS-CoV-2 replication. (A) Experimental scheme to establish A5494320 and A549ACE2/TMPRSS2 (ACE2plus). SARS-CoV-2 infection observed in ACE2plus (B) and 43.20 cells (C) at 48 hours post-infection. Spike-specific antibody was used to detect infected cells; 96-well plates were imaged with the ImageXpress, 4X magnification, 100uM scale bar. (D) Scanned images were analyzed using MetaXpress software to determine the infectivity (Spike+ cells were normalized to total cell number). In condition 1, 20,000 cells per well were seeded in a 96-well plate; 15,000 cells were seeded in condition 2. (E) ACE2plus shows comparable infectivity to Vero E6 cells while challenging with SARS-CoV-2 (WA1/2020) or icSARS-CoV2-mNG (WA1/2020). (F, G) Virus-containing supernatants from infected ACE2plus cells at 48 hours post-infection were collected and analyzed by RT-qPCR and plaque assay to measure the viral load and viral particles, their ability to form plaques. The data represent the mean (±SD) from two independent experiments.
The viral nucleoprotein ribonucleic acid (RNA) levels were quantified using real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR), for which supernatants were collected from cells treated with wild-type virus and icSARS-CoV-2-mNG at 48 hours post-infection (pi) and processed for RNA extraction.
Cell-cell fusion allows viruses to infect neighboring cells, for which the polybasic S1/S2 site of SARS-CoV-2 S is required, which also involves syncytium formation. The size of syncytia constitute a hallmark of coronavirus disease 2019 (COVID-19)-associated pathology; thus, it might be essential to understand the ability of syncytium formation between SARS-CoV-2 spike variants.
For this, the researchers first established a mCherry stable cell line using the ACE2plusC3 model. After sorting, over 98% of cells expressed mCherry protein; therefore, this cell model was used to observe the kinetic of S-mediated cell-cell fusion without coculturing with other cells. Next, they transfected an equal amount of indicated S plasmids into ACE2plusC3-mCherry cells for 24 hours.
The researchers also evaluated the efficacy of Camostat mesylate, EIDD-1931, and Remdesivir in inhibiting SARS-CoV-2 infection. Next, they investigated the efficacy of the furin inhibitor Decanoyl RVKR-CMK to determine the need for furin protease for syncytium formation in ACE2plucC3 cells. The cells were infected with WA1/2020 isolate at MOI 0.1, inoculated with different concentrations of Decanoyl-RVKR-CMK for 36 hours.
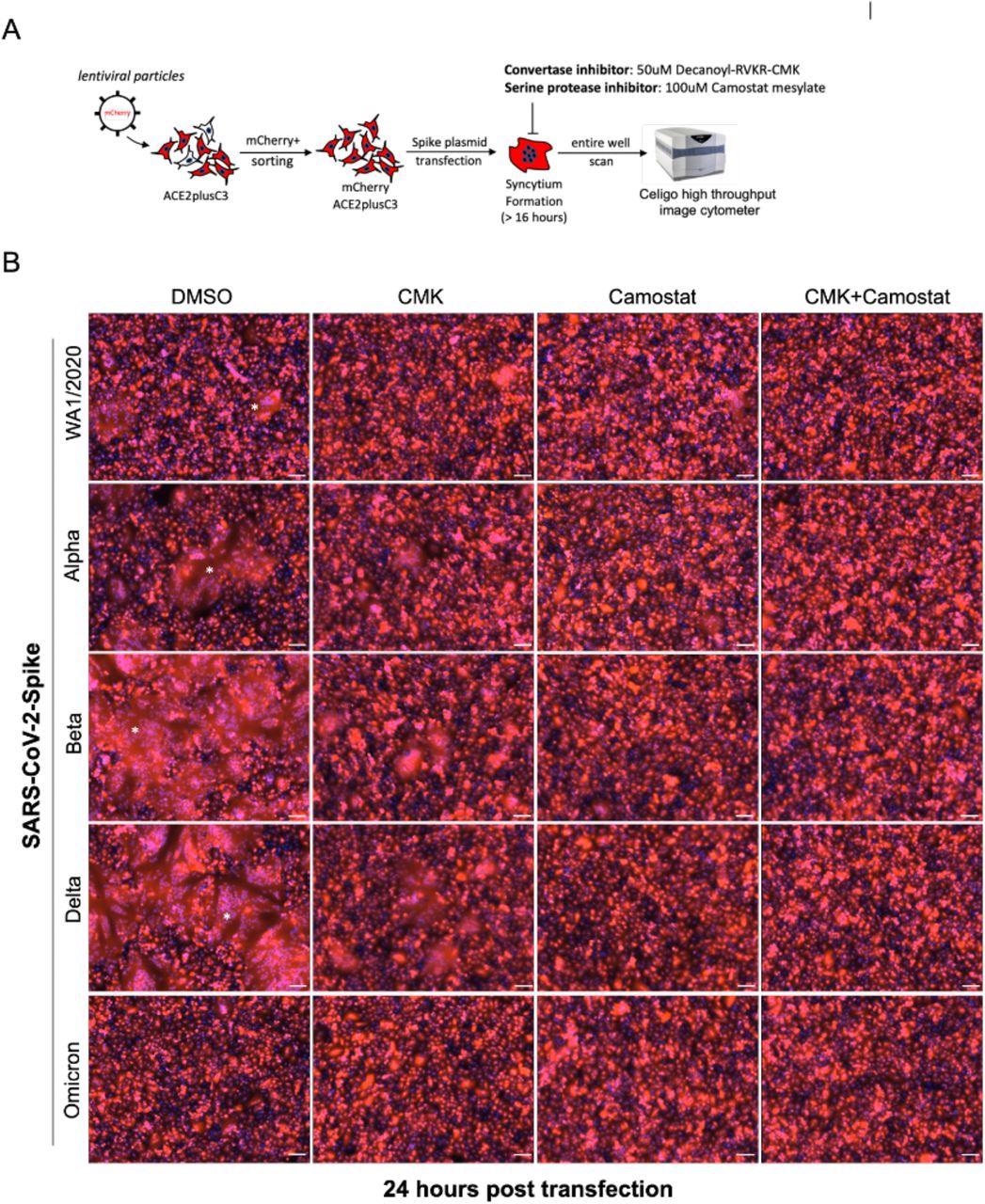
SARS-CoV-2-Spike-mediated syncytium formation in ACE2plusC3 cells. (A) Experimental scheme to observe syncytium formation. To visualize and capture whole well images, an entire 24-well plate was scanned (B) Representative images from two independent experiments. SARS-CoV-2 Spike plasmids were transfected into cells with or without protease inhibitor treatment. After 24 hours, cells were fixed and scanned using Celigo Image Cytometer. mCherry and DAPI images were merged by Celigo Software. 100uM scale bar. Star shows the area of the syncytium.
Study findings
Even during low multiplicity of infection (MOIs) of 0.05 and 0.1, ACE2plus cells showed higher infectivity than the parental 43.20 cells. MOI refers to the number of virions added per cell during infection.
As compared to ACE2plus cells, Vero E6 cells showed 1.75-fold higher infectivity when treated with wild-type virus and about 1.2 fold higher infectivity when treated with icSARS-CoV-2-mNG. Altogether, the study results showed comparable efficacy of both ACE2plus and Vero E6 cells for modeling SARS-CoV-2 infections.
The study results also demonstrated that the single-cell derived subclone of ACE2plusC3 was highly susceptible to SARS-CoV-2 infection and showed higher ACE2 and TMPRSS2 messenger RNA (mRNA) expression levels than commercially used Calu3 cells. Further, the ACE2 receptors on the surface of ACE2plusC3 cells were recognized by SARS-CoV-2 receptor-binding domain (RBD) protein, thus supporting the use of ACE2plusC3 for SARS-CoV-2 257 lentiviral infection assays.
Camostat mesylate, EIDD-1931, and Remdesivir exhibited potent antiviral effects with half-maximal inhibitory concentration (IC50) of 59.98 uM, 0.84 uM, and 0.14 uM, respectively. These results demonstrate that the ACE2plucC3 cell model could be used for the evaluation of antiviral drugs.
Multinucleated syncytia is a common clinical feature of severe COVID-19 induced in ACE2plusC3 cells either by a viral infection or by overexpressing the spike proteins of SARS-CoV-2 variants. The SARS-CoV-2 Delta and Beta variant spike proteins induced giant syncytia formation of greater than 300uM, of multinucleated enlarged cells, as compared with WA1/2020 or alpha S.
The syncytia formation was reduced upon adding furin protease inhibitor, decanoyl RVKR-CMK, or Camostat inhibitors to transfected cells. Further, the SARS-CoV-2 Omicron variant spike protein poorly induced cell fusion in ACE2plus mCherry cells, thereby indicating that the ACE2plusC2 model can be used to study S-mediated cell fusion.
Conclusions
The present study provides data on SARS-CoV-2 production, pseudotyped infection, spike-mediated cell-to-cell fusion, and antiviral tests to highlight the importance of the human lung-based cell model for studying SARS-CoV-2 and its continually emerging variants.
In particular, the A549ACE2PlusC3 cell model emerged as an alternative and improved model for in vitro disease modeling, which is useful for COVID-19 drug discovery and host-viral interaction studies. Overall, the study showed a robust cell model based on human lung cells, which could become a valuable resource for the research community studying SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Chang, C., Parsi, K. M., Somasundaran, M., et al. (2021). Cell culture model system utilizing engineered A549 cells to express high levels of ACE2 and TMPRSS2 for investigating SARS-CoV-2 infection and antivirals. bioRxiv. doi:10.1101/2021.12.31.474593. https://www.biorxiv.org/content/10.1101/2021.12.31.474593v1.
- Peer reviewed and published scientific report.
Chang, Ching-Wen, Krishna Mohan Parsi, Mohan Somasundaran, Emma Vanderleeden, Ping Liu, John Cruz, Alyssa Cousineau, Robert W. Finberg, and Evelyn A. Kurt-Jones. 2022. “A Newly Engineered A549 Cell Line Expressing ACE2 and TMPRSS2 Is Highly Permissive to SARS-CoV-2, Including the Delta and Omicron Variants.” Viruses 14 (7): 1369. https://doi.org/10.3390/v14071369. https://www.mdpi.com/1999-4915/14/7/1369.