
 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Several SARS-CoV-2 variants have emerged since the beginning of the current coronavirus disease 2019 (COVID-19) pandemic, with the Omicron variant being the most recently identified variant of concern (VOC). Shortly after its emergence, the SARS-CoV-2 Omicron variant quickly spread across the world leading to a sharp rise in the number of COVID-19 cases.
Several studies have shown that the high number of mutations in the spike protein of the Omicron variant confers immune escape properties including evasion of polyclonal antibody responses. Widespread reinfections and vaccine breakthrough infections by the Omicron variant have been reported globally.
About the study
In the present study, a team of researchers analyzed and compared the growth of the SARS-CoV-2 Omicron variant with the Delta variant and other variants in different cell lines including Vero E6 cells overexpressing human angiotensin-converting enzyme-2 (ACE2) and transmembrane protease, serine-2 (TMPRSS2) [Vero-AT cells], Calu-3 cells, and primary cultures of human nasal epithelial cells (hNECs). Furthermore, the authors investigated whether the Omicron variant differed from other VOCs in utilizing ACE2 and TMPRSS2 for entry into cells and whether its high number of mutations is associated with any biological changes in the hosts.
Study findings
The authors noted that both the Delta and Omicron variants produced plaques in Vero-AT cells; however, the plaques formed by the Omicron variant were smaller and less distinct. In a mixed competition assay involving equal inoculates of both Delta and Omicron variants, the authors found that the Omicron variant outcompeted the Delta variant in hNECs, while in Vero-AT cells, both variants showed similar growth. However, in Calu-3 cells, the Delta variant outcompeted the Omicron variant and remained dominant.
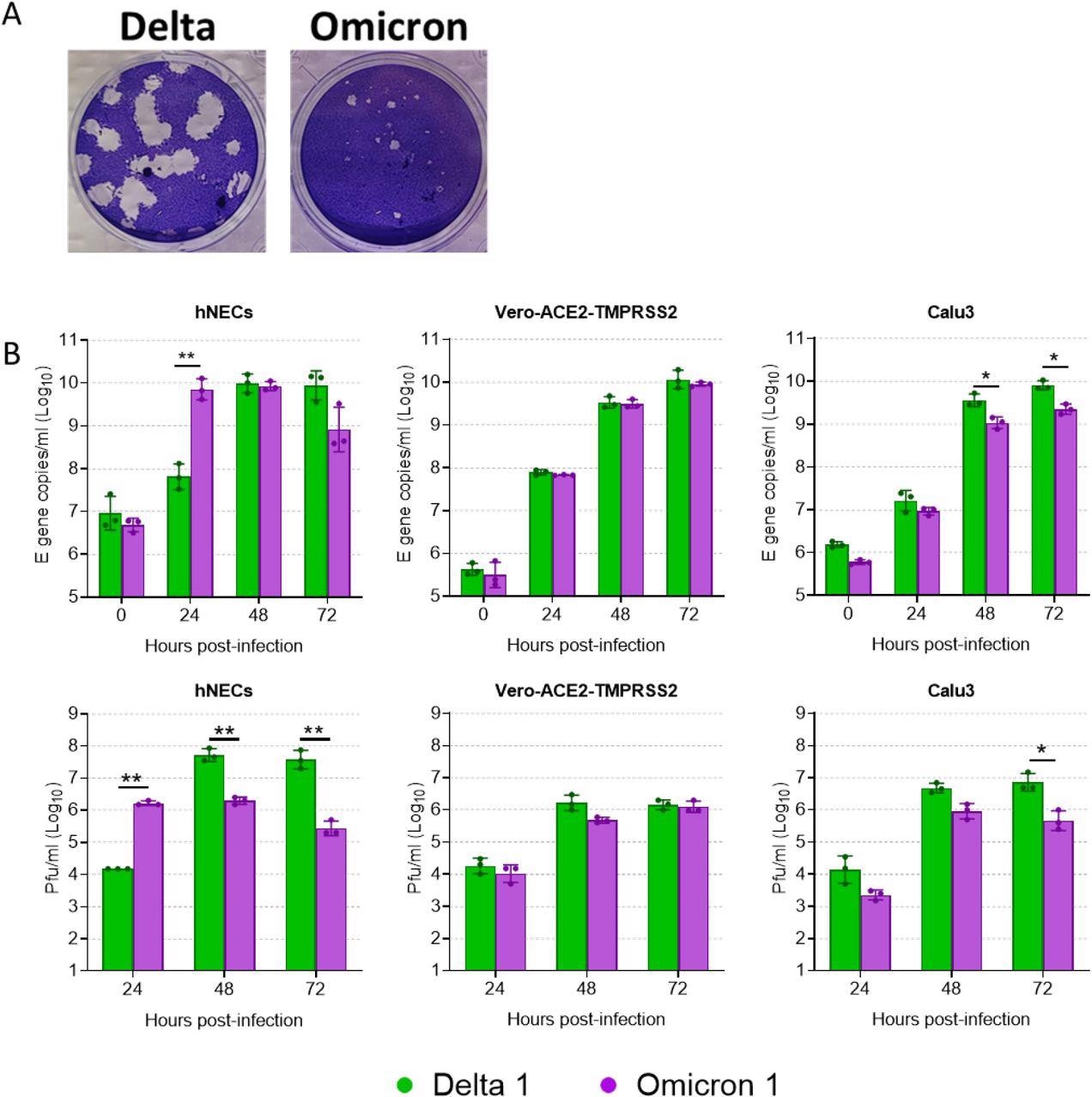
Comparative replication kinetics of Omicron and Delta. (A) Virus isolates were propagated on Vero-ACE2-TMPRSS2 (VAT) cells and plaqued on VAT cells for 3 days before visualizing by crystal violet staining. (B) Comparative replication kinetics of SARS-CoV-2 Omicron and Delta variants in vitro and ex vivo. Primary human nasal epithelial cultures (hNECs) were inoculated with Delta or Omicron variants at 0.1 pfu/cell, and Vero-ACE2-TMPRSS2 (Vero-AT) and Calu-3 cells were inoculated at 0.001 pfu/cell. Infections were performed in triplicate wells and back titration confirmed equal inputs of the variants by E gene copies measured by RT-qPCR and infectious units measured by plaque assay. Bars show individual replicate titers with the standard deviation. Statistical differences were measured by ANOVA on log-transformed data. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001
The researchers investigated the portal of entry of Omicron spike-containing lentiviral pseudotypes in 293T cells that expressed different canonical human coronavirus receptors including ACE2, aminopeptidase N (APN), and dipeptidyl peptidase-4 (DPP4). It was observed that the Omicron variant entered the cells efficiently when ACE2 was overexpressed, which suggests that ACE2 remains the preferred protein receptor and portal of entry for the SARS-CoV-2 Omicron variant. The full spike protein of the Omicron variant was found to bind to human ACE2 to a greater extent than that of the Alpha or Delta variant.
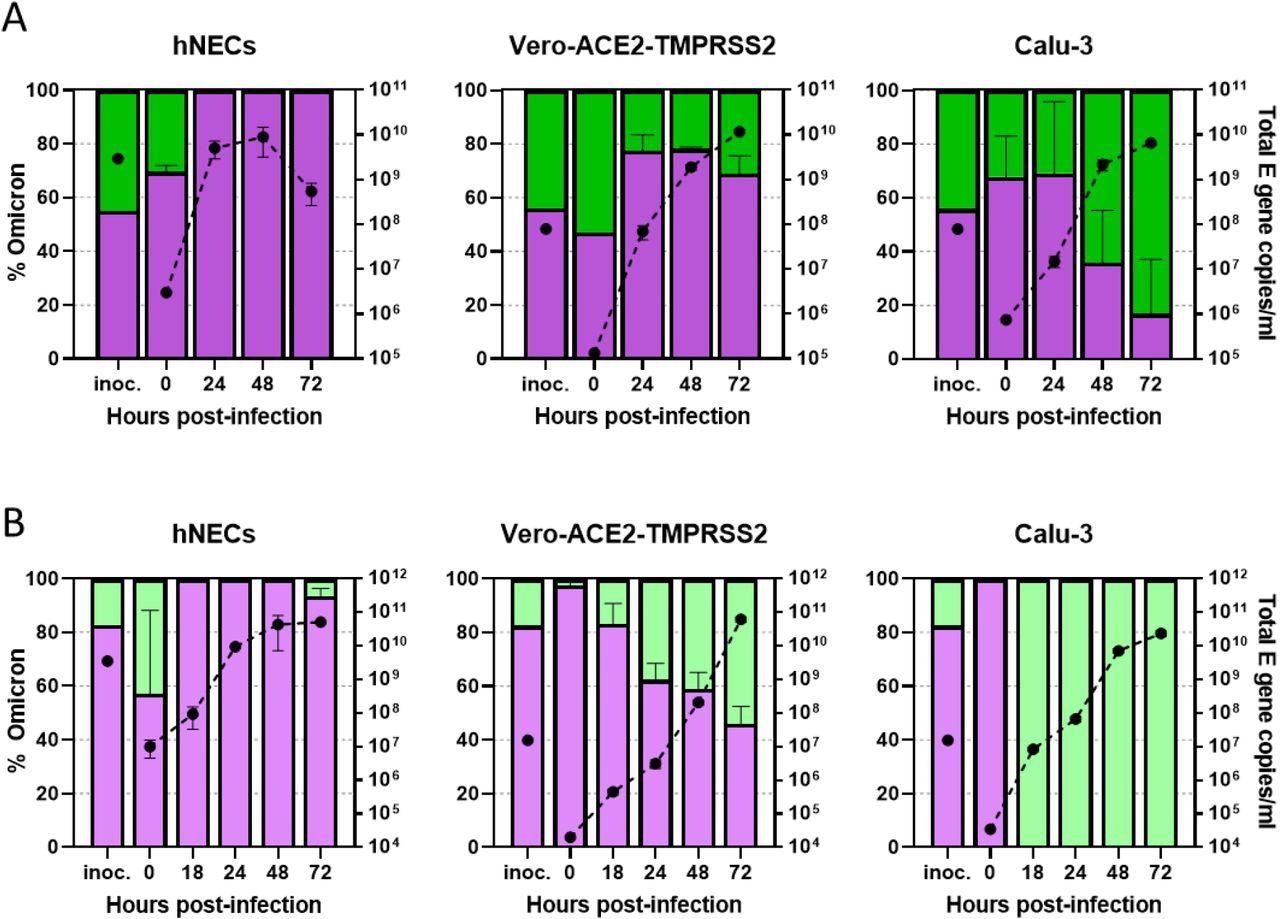
Competition assays between Delta and Omicron isolates. (A,B) Competition assay between SARS-CoV-2 Omicron (Purple) and Delta (Green) isolates in vitro and ex vivo. A probe-based RT-qPCR assay was developed to measure the proportion of Omicron and Delta present in mixed samples (see Supplementary Figure S1). The proportion of variant-specific signal is shown with the SD of 3 replicates. (A) Equal mixtures or (B) an 8:2 ratio of Omicron and Delta isolates were inoculated onto hNECs at a final multiplicity of 0.1 pfu/cell and onto Vero-AT and Calu-3 cells at 0.001 pfu/cell. Daily harvests were measured for total viral load by E gene qRT-PCR (black dots) and by the variant-specific RT-qPCR to give variant proportions. Mean + s.d. of 3 replicates shown. (A) Shows competition assay specifically between BA.1 (Spike A701V) and a B.1.617.2 isolate while (B) shows competition between a different BA.1 isolate (lacking A701V) an AY.4.2 isolate.
The findings of the current study showed that the Omicron variant uses ACE2 of several species to enter cells to a greater extent than any other SARS-CoV-2 variant. SARS-CoV-2 variants induce the formation of syncytia, which require the spike protein to be pre-cleaved at the S1/S2 junction. Although the Omicron variant contains mutations that enhance cleavage, the researchers observed attenuated syncytia formation. The authors noted that the Omicron variant can use the endosomal pathway of entry while avoiding innate factors like interferon-induced transmembrane (IFITM) proteins.
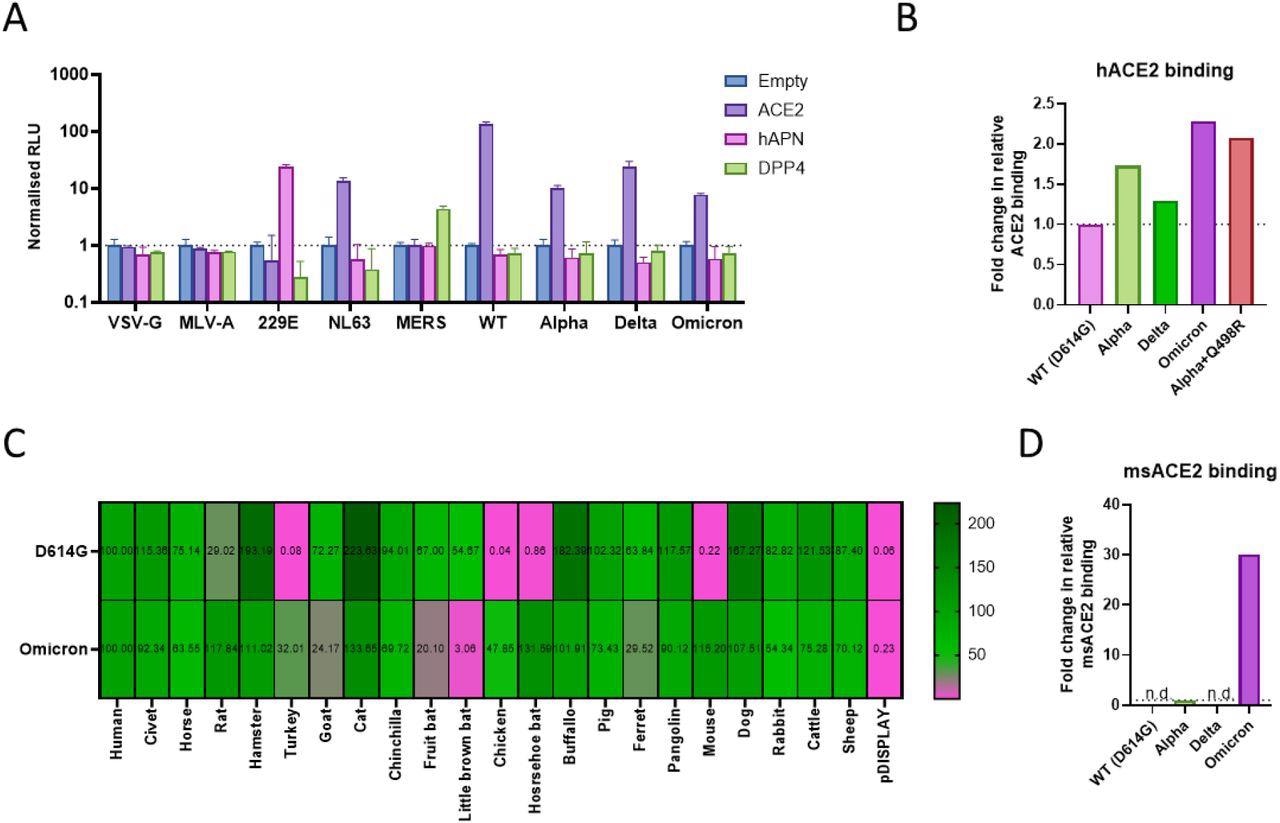
Use of common coronavirus protein receptors and receptor species specificity for SARS-CoV-2 variant entry. (A) Entry of pseudotyped lentiviruses expressing different viral glycoproteins was assessed into 293Ts transfected with different coronavirus protein receptors. 229E and MERS Spike protein pseudotypes were used as controls for hAPN and DPP4 mediated entry. All assays were performed in triplicate and are plotted as mean + s.d. (B, D) HEK 293T cells expressing whole SARS-CoV-2 Spike were incubated with recombinant (B) hACE2-Fc(IgG) for 30 minutes or (D) mouse ACE2-Fc(IgG) overnight, washed and incubated with Goat Anti-Human IgG Fc (DyLight® 650) secondary for 30 minutes, before washing and analysis by flow cytometry. Relative differences in median fluorescence intensity are shown. ‘n.d.’ indicates not detected. (C) Receptor usage was screened using pseudoviruses expressing the indicated Spike proteins into BHK-21 cells expressing the indicated ACE2 protein. Viral entry was measured by assaying luciferase activity (RLU) using the BrightGlo reagent (Promega).
Conclusions
The observations made in the present study revealed that the SARS-CoV-2 Omicron variant exhibits significant biological changes when compared to other variants. The researchers highlighted the rapid and increased replication of the Omicron variant in the primary cultures of hNECs that outcompete the Delta variant and any other SARS-CoV-2 variant.
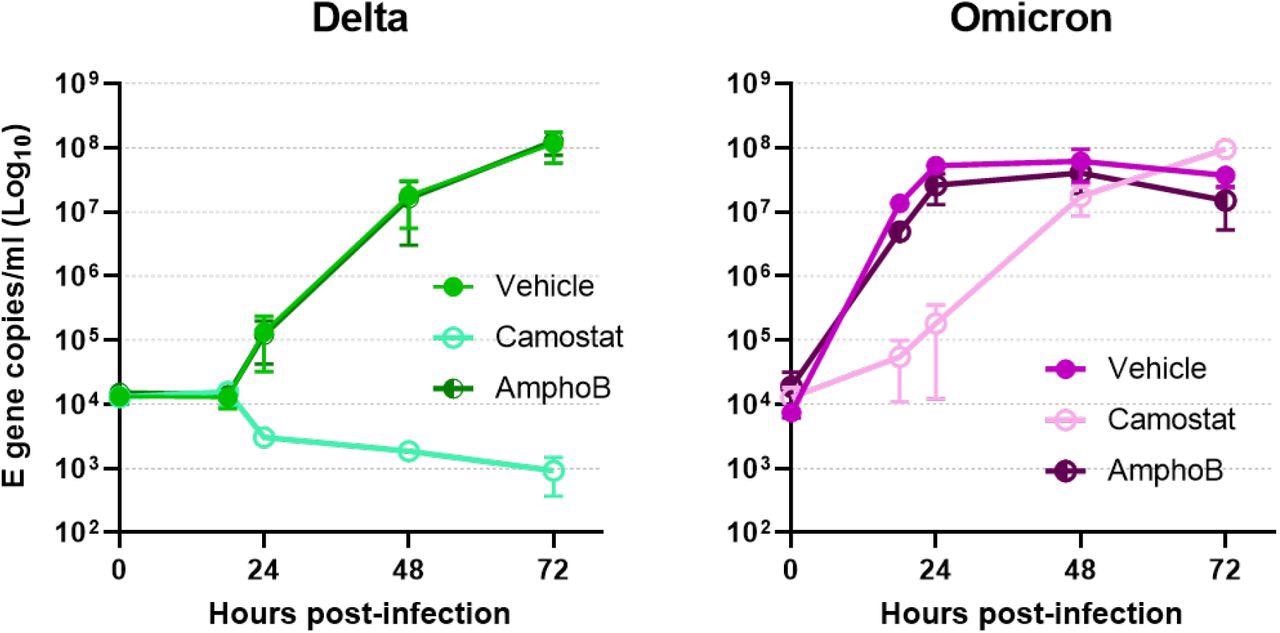
Cell entry mechanism of live Omicron virus in primary cells. Comparative replication kinetics of SARS-CoV-2 Omicron and Delta variants in primary human nasal epithelial cultures (hNECs) inoculated with Delta or Omicron variants at 0.1 pfu/cell. Cells were pretreated both basolaterally and apically with either with no drug, or with 50 µM of Camostat or Amphotericin B for 2 hours prior to infection. 50µM of Camostat or Amphotericin B remained in the basolateral media for the course of the experiment. Infections were performed in triplicate wells and back titration confirmed equal inputs of the variants by E gene copies measured by RT-qPCR and infectious units measured by plaque assay. Plotted as mean ± s.d.
The authors of the current study also proposed that the rapid replication of Omicron could be due to its reduced cellular tropism. Earlier variants required TMPRSS2 protease for cell entry including the Alpha and Delta VOCs; however, the current study demonstrated that the Omicron variant uses an endosomal pathway for entry that increases the number of cell types it can infect. This switch of entry pathway could explain the increase in transmissibility of the Omicron variant.
The researchers also reported a reduced propensity for syncytia formation by the Omicron variant, which could explain the reduced severity of COVID-19 in the infected population when compared to other SARS-CoV-2 variants. This decrease in disease severity, however, does not lower the healthcare burden caused by the rapid surge in the number of COVID-19 cases and therefore highlights the need for improvising existing vaccination strategies to curb the spread of the virus in the future.
The finding that the Omicron variant can infect cells from different species more efficiently than other SARS-CoV-2 variants indicates the ability of this virus to establish a more potent animal reservoir, thus limiting the chances of SARS-CoV-2 from being completely eradicated.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.