Every time a new variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerges, the question arises, “is it going to swap the previously circulating SARS-CoV-2 strain around the world and cause more severe disease?”
Study: No evidence for increased cell entry or antibody evasion by Delta sublineage AY.4.2. Image Credit: Corona Borealis Studio / Shutterstock
Background
As a consequence of the high mutation rate of SARS-CoV-2, several variants have emerged since the beginning of the coronavirus disease 19 (COVID-19) pandemic. Mutations in the viral spike S protein are of particular importance as the S protein drives the host cell entry and acts as the neutralizing target for the protective antibodies. Therefore, the mutations in the S protein can modulate viral transmissibility and pathogen capacity.
To date, the World Health Organization (WHO) has classified five variants of concern, Alpha (B.1.1.7 lineage), Beta (B.1.351 lineage), Gamma (P.1 lineage), Delta (B.1.617.2 lineage), and Omicron (B.1.1.529 lineage). A lineage refers to a genetically closely related group of virus variants derived from a common ancestor. For example, the Delta variant B.1.617.2 lineage, until recently the dominant variant around the world, has certain factors attributable to its advantage, including efficient host cell entry and improved antibody escape potential.
Several sublineages, harboring additional mutations in the spike S protein, have branched off from the parental B.1.617.2 Delta lineage. The SARS-CoV-2 lineage AY.4.2, representing a sublineage of the Delta variant parental lineage B.1.617.2, is currently behind 2.1–19.4% of new cases in the UK. However, no information is available regarding its disease-causing and antibody-neutralization ability compared to the parental virus lineage.
Study methodology
The mutations present in the S protein of the SARS-CoV-2 lineage AY.4.2 were analyzed.
Using a pseudotype virus approach, the team compared the ability of AY.4.2 and B.1.617.2 S protein to drive entry into target cells. Another lineage, B.1 that circulated in the pandemic's early phase was also included in the comparison.
Further, the team evaluated the neutralization efficiency of five clinical therapeutic antibodies against the ribosomal binding domain (RBD) of AY.4.2 S protein compared to that of B.1.617.2 and B.1 lineages.
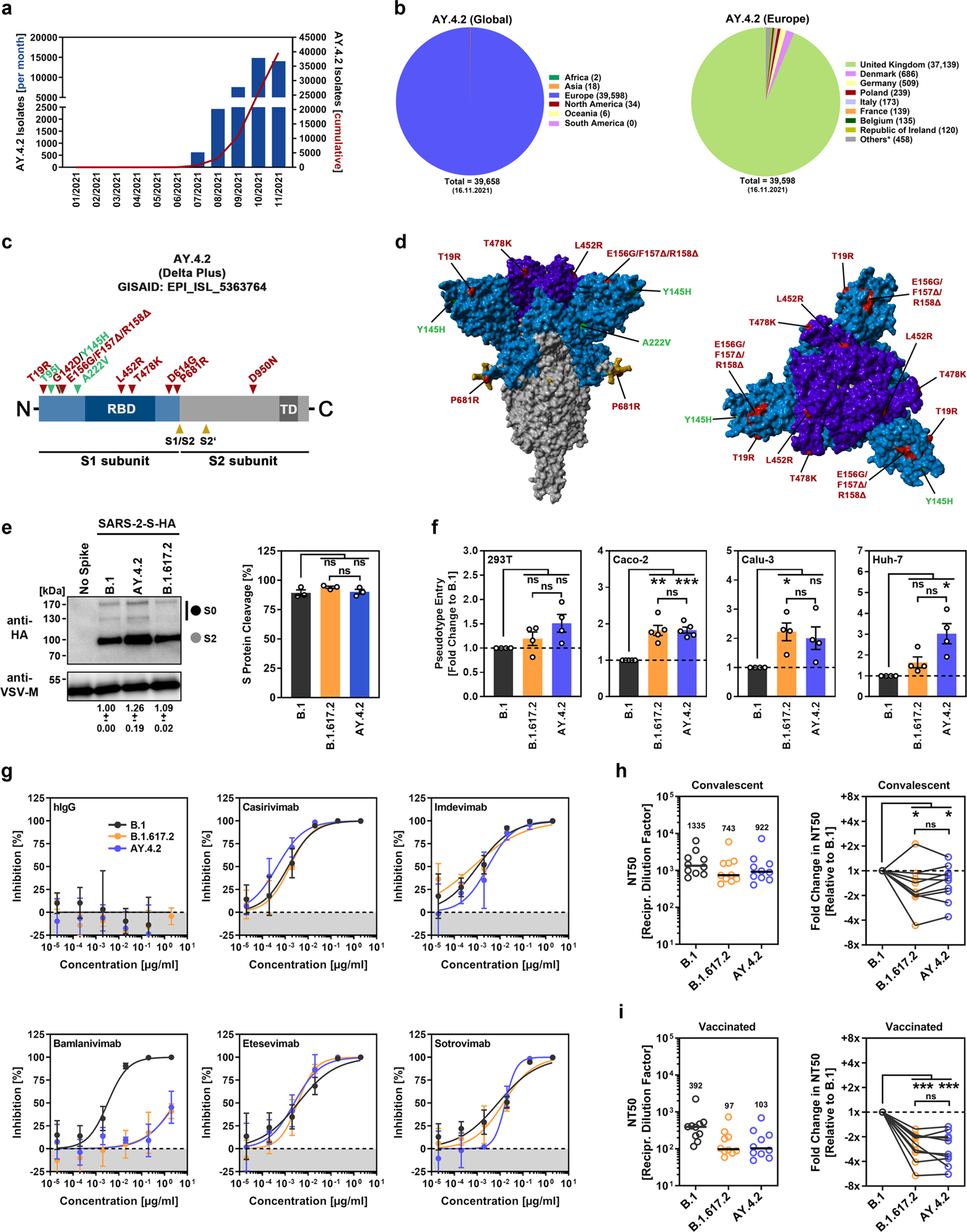
a Monthly and cumulative numbers of globally reported AY.4.2 isolates. b Distribution of reported AY.4.2 isolates at the global (left) and European (right) levels. Numerical values in brackets indicate the number of isolates per country (* = twenty countries with < 100 isolates). c Schematic illustration of the SARS-CoV-2 spike protein in which the locations of functional domains (RBD, receptor binding domain; TD, transmembrane domain) and cleavage sites (S1/S2 and S2’) are highlighted. Mutations found in the spike protein of B.1.617.2 (Delta variant, EPI_ISL_1921353) are highlighted in red, while the additional mutations found in the Delta sublineage AY.4.2 (EPI_ISL_5633764) are highlighted in green. d Location of the amino acid changes in the context of the trimeric spike protein. e Pseudotyped particles bearing the indicated S proteins (equipped with a C-terminal HA epitope tag) were subjected to immunoblot analysis to analyze S protein incorporation and cleavage. S proteins and VSV-M (loading control) were detected using anti-HA and anti-VSV-M antibodies, respectively, in combination with a peroxidase-conjugated anti-mouse secondary antibody. The results from a single experiment are presented, and the results were confirmed in two additional experiments. For quantification of S protein incorporation, in all experiments, the S protein signals were first normalized against the corresponding VSV-M signals, and further incorporation of B.1 S protein was set as 1 (data represent the mean ± standard deviation, SD). For quantification of S protein cleavage, total S protein signals (bands representing unprocessed [S0] and processed [S2] S protein) for each S protein were set as 100%, and the respective proportions of S0 and S2 were calculated. Statistical significance was assessed by two-tailed Student’s t test with Welch’s correction; p > 0.05, not significant [ns]). f Four different human cell lines were inoculated with pseudotyped particles bearing the indicated spike proteins. At 16–18 h postinoculation, particle entry efficiency was analyzed by measuring the activity of virus-encoded luciferase in cell lysates. Presented are the average (mean) data from 4–5 independent experiments (each performed with four technical replicates) for which particle entry driven by the B.1 spike protein was set as 1. Error bars indicate the standard error of the mean. Statistical significance was assessed by two-tailed Student’s t-test with Welch’s correction (p > 0.05, ns; p ≤ 0.05, *; p ≤ 0.01, **; p ≤ 0.001, ***; please see also Supplemental information, Fig. S1a). g Pseudotyped vectors bearing the indicated spike proteins were incubated (30 min, 37 °C) in the presence of different concentrations of monoclonal antibody or medium alone (control) before being added to Vero cells. Vector entry efficiency was analyzed at 16–18 h postinoculation and normalized against the respective control (set as 0% inhibition). Presented are the average (mean) data for a single experiment (with four technical replicates). The data were confirmed in a separate experiment. Error bars indicate the SD. Curves were calculated using a nonlinear regression model (variable slope). h Pseudotyped vectors bearing the indicated spike proteins were incubated (30 min, 37 °C) in the presence of different dilutions of convalescent plasma or only medium (control) before being added to Vero cells. Vector entry efficiency was analyzed at 16–18 h postinoculation and normalized to the respective control (set as 0% inhibition, please see Supplemental information, Fig. S1b for individual data). Furthermore, the plasma dilution that causes a reduction of 50% in vector entry (neutralizing titer 50, NT50) was calculated. Presented are the combined data for 10 convalescent plasma (each analyzed in four technical replicates). Black lines and numerical values indicate the median NT50. In addition, the data were normalized to reflect the relative change in neutralization sensitivity with the neutralization of B.1 spike serving as reference (set as 1, identical plasma are connected by lines). Statistical significance was assessed by Kruskal–Wallis analysis with Dunn’s multiple comparison test (p > 0.05, ns; p ≤ 0.05, *; p ≤ 0.001, ***). i The experiment was performed as described in (h), but serum from vaccinated individuals (BNT162b2/BNT162b2, n = 10) was analyzed.
Study findings
Initial mutational analysis revealed that the S protein of AY.4.2 encompassed characteristic amino acid mutations (L452R and T478K) of B.1.617.2 in RBD that have been shown to reduce the efficacy of therapeutic antibodies and, along with other mutations found in an antigenic supersite within the N-terminal domain (G142D, E156D, F157Δ, R158Δ), may bestow the virus with an escape mechanism from neutralization by self-antibodies, induced upon infection or vaccination.
Another S protein mutation, P681R, reported to be associated with coronavirus disease 2019 (COVID-19) pathogenesis via augmenting cell-to-cell fusion, was also found in AY.4.2 S protein.
Compared to the previously circulating SARS-CoV-2 B.1 lineage, the S proteins of both AY.4.2 and B.1.617.2 enhanced the viral entry by nearly 2-fold into the human lung (Calu-3) and colon-derived cells (Caco-2). In addition, the entry efficiency of AY.4.2 and B.1.617.2 into the kidney-derived 293T cell line was equal to that of B.1. Except for a moderately more efficient (~2-fold, but not statistically significant) entry into the human liver Huh-7 cell line by the AY.4.2 S protein, the team did not observe any differences in cell entry efficiency between AY.4.2 and B.1.617.2 S proteins.
Regarding the therapeutic antibodies, four antibodies-Casirivimab, Etesevimab, Imdevimab, and Sotrovimab-were found to efficiently neutralize the S proteins of B.1, B.1.617.2, and AY.4.2. The antibody Bamlanivimab remained mostly ineffective against both B.1.617.2 and AY.4.2. The team speculates that it could be due to the presence of L452R mutation in both S proteins.
Concerning neutralization by self-antibodies, S proteins of both AY.4.2 and B.1.617.2 were less efficiently neutralized by either convalescent plasma (median 1.3- and 1.6-fold reduction for AY.4.2 and B.1.617.2, respectively) or sera from BNT162b2-vaccinated individuals (median 2.8- and 2.3-fold reduction for AY.4.2 and B.1.617.2, respectively) compared to the S protein of B.1.
Conclusions
The study sheds light on the nature of the S protein of AY.4.2 virus lineage. Furthermore, it reveals no appreciable differences between the pathogenic potential and antibody susceptibility of AY.4.2 and B.1.617.2 virus lineages. The finding thus implies that the current treatment options and vaccination will be equally effective against both the variants.