With more than 360 million cases worldwide and over 5.59 million deaths that have been caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, it has become increasingly urgent to identify markers that can help determine which infected patients are at a high risk of complications due to the coronavirus disease 2019 (COVID-19).
Study: EFCAB4B (CRACR2A) Genetic Variants Associated With COVID-19 Fatality. Image Credit: CROCOTHERY / Shutterstock.com

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
Though generally a mild to moderate illness, COVID-19 progresses in a sizable minority of patients to severe or even fatal multi-organ failure and acute respiratory distress syndrome (ARDS). The reason for this difference in prognosis is thought to be at least partly due to cardiovascular complications such as abnormal clot formation and endothelial inflammation-causing injury of the microvasculature.
Endothelial damage by SARS-CoV-2 can activate endothelial cells that lead to the release of factors that promote clotting. These clotting factors are found at higher levels in severe COVID-19 patients and are therefore regarded as markers of increased mortality. The cytokine storm that has garnered much publicity in this setting is also considered to be an indicator of critical illness, as these cytokines are traceable to endothelial cell activation.
Cytokines are stored within the endothelial cells in Weibel-Palade bodies (WPBs) and their release is regulated to allow for events like hemostasis, vascular growth, vascular tone, and inflammation. Such release, or degranulation, occurs during vascular injury, subsequently leading to platelet plug formation, leukocyte chemotaxis, and endothelial cell migration. A positive feedback loop ensures that chemokines also stored in the WPBs activate other endothelial cells to amplify the process.
About the study
The current study examines the hypothesis that the degranulation of endothelial cells is partly responsible for the clotting and inflammation in severe COVID-19 at the molecular level. This could explain why only some patients appear to develop severe illnesses.
The researchers of the current study have previously identified a novel Rab GTPase (Rab46: CRACR2A-L) that is required to anchor a subpopulation of WPBs within the cell when histamine is released. This prevents the unnecessary degranulation of endothelial cells.
Mutations in Rab46 impact the release of WPB cargo, thereby indicating its regulatory role. There are two functional isoforms of the EFCAB4B gene transcript, of which Rab46 alone is expressed in endothelial cells, whereas CRACR2A is expressed in T-cells and regulates cell signaling and calcium influx.
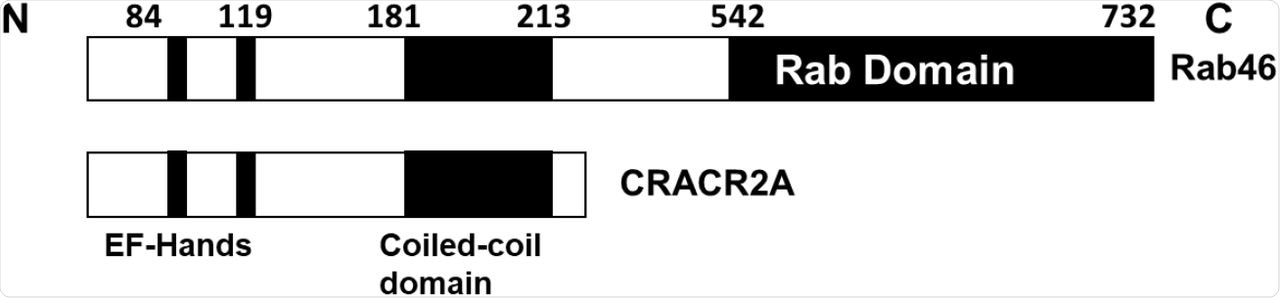
Schematics of the functional domains in the EFCAB4B isoforms Rab46 and CRACR2A.
Previously, the researchers found that mutations in both alleles of this gene cause impaired T-cell signaling and lowered immune function with poor cytokine production. Similarly, among convalescent patients following mild COVID-19, CRACR2A levels were reduced, as inflammation gave place to repair and regenerative processes.
Using an approach called candidate gene association study, the scientists attempted to identify links between mutations in this gene and mortality due to COVID-19. The data used in the current study was acquired from the United Kingdom Biobank stored data that was collected before the rollout of the vaccination program.
Study findings
Among the approximately 1,260 COVID-19 patients included in the current study, there was a 15% mortality rate.
The researchers looked at those SNPs that alter the EFCAB4B gene translation into its gene products. While missense mutations were found to occur with three SNPs in both isoforms, these three were also linked to a fatal outcome from COVID-19, with the odds of death being increased by 40% to 50%.
These missense mutations caused amino acid substitutions in both isoforms. One was a shift from arginine to glycine, another alanine to threonine, and the third was histidine to glutamine.
Two of the identified mutations are in functionally important and highly conserved positions in the domains of both isoforms (98% and 99% conserved loci, respectively). Comparatively, the arginine-glycine mutations are located in a structurally undefined region, but at an 80% conserved locus.
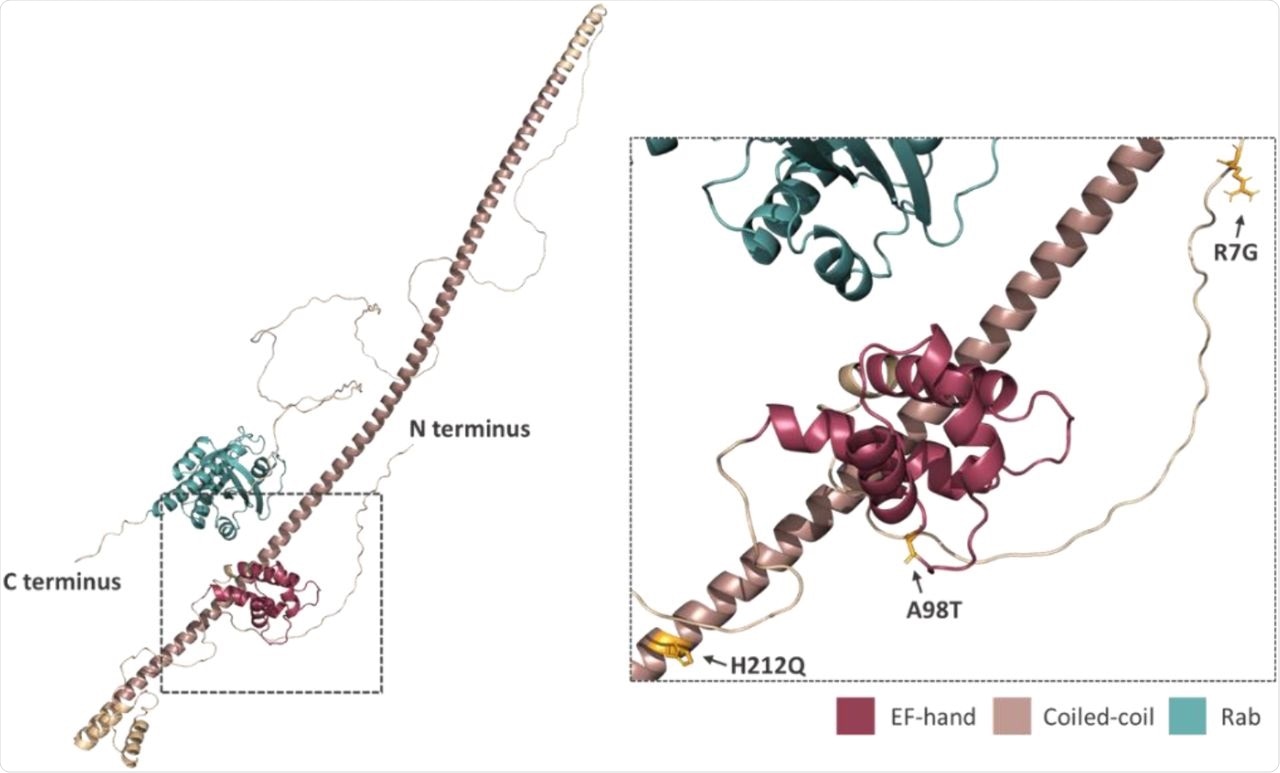
Prediction of the position and orientation of the three missense SNPs in an AlphaFold model of full-length Rab46 structure. The box indicated by the dotted line in the full-length structure is enlarged in order to demonstrate the position and orientation of the side chains of the three missense SNPs: R7G; A98T; H212Q.
This data indicates how important these residues are to Rab46 function. Structural analysis of this protein suggests outward-facing side chains.
For the A98T mutation, small changes were found to occur in the binding domain, with a wild-type structure of aspartate/alanine/aspartate. The changes caused the two aspartate residues to overly one another, which could potentially affect calcium ion coordination and subsequently impact protein conformation. While tolerated under normal conditions, it is possible that disease or stress could induce a harmful change in protein function, accounting for its association with COVID-19 mortality.
The histidine-glutamine substitution occurs in a coiled-coil region that typically enhances protein-protein interactions. The positive charge on the histidine imidazole ring could therefore be key for metal coordination to strengthen an oligomerized protein complex. The outward-facing nature of this residue would indicate its importance in interactions between molecules, rather than stabilizing the helix.
Implications
No missense mutations were linked to non-fatal COVID-19; however, three identified mutations were significantly associated with fatal disease following SARS-CoV-2 infection. This observation appears to rule out increased susceptibility to infection with these SNPs but increased risk of fatal illness.
Other alanine-threonine mutations in amyloid and similar pathological proteins show a phenomenon called antagonistic pleiotropism. This refers to a type of mutation that is beneficial in the young population but becomes detrimental over time.
“Since Rab46 is a Rab GTPase that regulates both endothelial cell secretion and T-cell signalling, these missense variations may play a role in the molecular mechanisms underlying the thrombotic and inflammatory characteristics observed in patients with severe COVID-19 outcomes.”
Earlier studies have indicated a role for Rab46 and CRACR2A in the immune response to COVID-19, thereby suggesting that these proteins must be regulated in order to ensure a favorable course of illness with recovery. Further structural studies will be required to fully understand the significance of these mutations.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.