In a study conducted at the Florida International University, USA, scientists have investigated the impact of spike receptor-binding domain (RBD) mutations of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) omicron variant on its interaction with human angiotensin-converting enzyme 2 (ACE2).
Study: Significance of the RBD mutations in the SARS-CoV-2 Omicron: from spike opening to antibody escape and cell attachment. Image Credit: Fit Ztudio/Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Soon after its emergence in South Africa in November 2021, the omicron variant of SARS-CoV-2 has caused a rapid increase in new coronavirus disease 2019 (COVID-19) cases worldwide. Compared to previously circulating variants, the spike protein of the omicron variant contains more than 30 mutations, including 15 mutations specifically in the spike RBD. This makes the variant more transmissible and antibody resistant than previously circulating variants, including delta.
Currently, cases with omicron infection have been detected in more than 90 countries across the globe. In the USA, the omicron variant replaced the delta as the dominant variant within three weeks after its detection. To understand the impact of the omicron variant on the epidemiology of delta variant, it is essential to investigate how omicron spike mutations influence its transmissibility and immune escape ability. In this context, studies have shown that immunity induced by omicron infection is sufficient to neutralize delta infection.
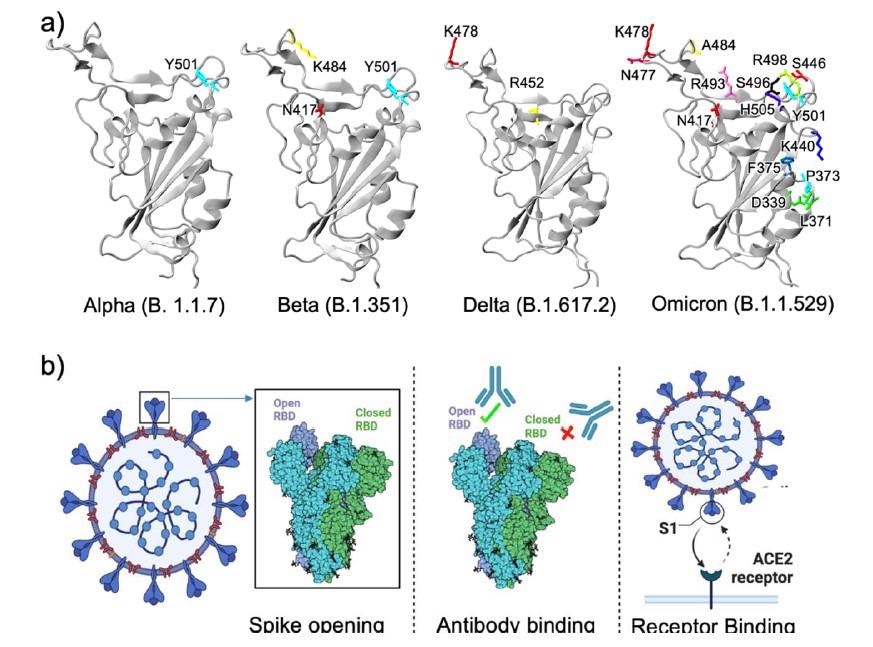
RBD structures of different variants. While Alpha, Beta, and Delta variants have less than three mutations in the RBD, Omicron has a remarkably large number of mutations. b) Three different ways RBD mutations may contribute to the high transmissibility of a variant (Created with BioRender.com).
In the current study, the scientists have conducted computational analyses to investigate the impact of omicron RBD mutations on ACE2 binding and antibody escape abilities.
The scientists performed molecular dynamics simulations of the spike RBD of wild-type SARS-CoV-2 and delta and omicron variants.
Impact of mutations on RBD opening
The interaction between spike RBD and ACE2 requires RBD opening from the closed-form spike trimer. The inter-domain hydrogen bonds that hold together three RBDs in the closed-form trimer are needed to break during RBD opening.
The analysis conducted in the study revealed that the majority of wild-type hydrogen bonds are present in the omicron variant. However, one primary hydrogen bond (Y505(A)-F374(B)) was lost in omicron because of the Y505H mutation. In addition, minor hydrogen bonding with multiple residues was lost due to polar to hydrophobic mutation S371L. The formation of new hydrogen bonds compensated the abrogation of wild-type hydrogen bonding in omicron. These new interactions facilitated the omicron RBD opening from the closed-form trimer.
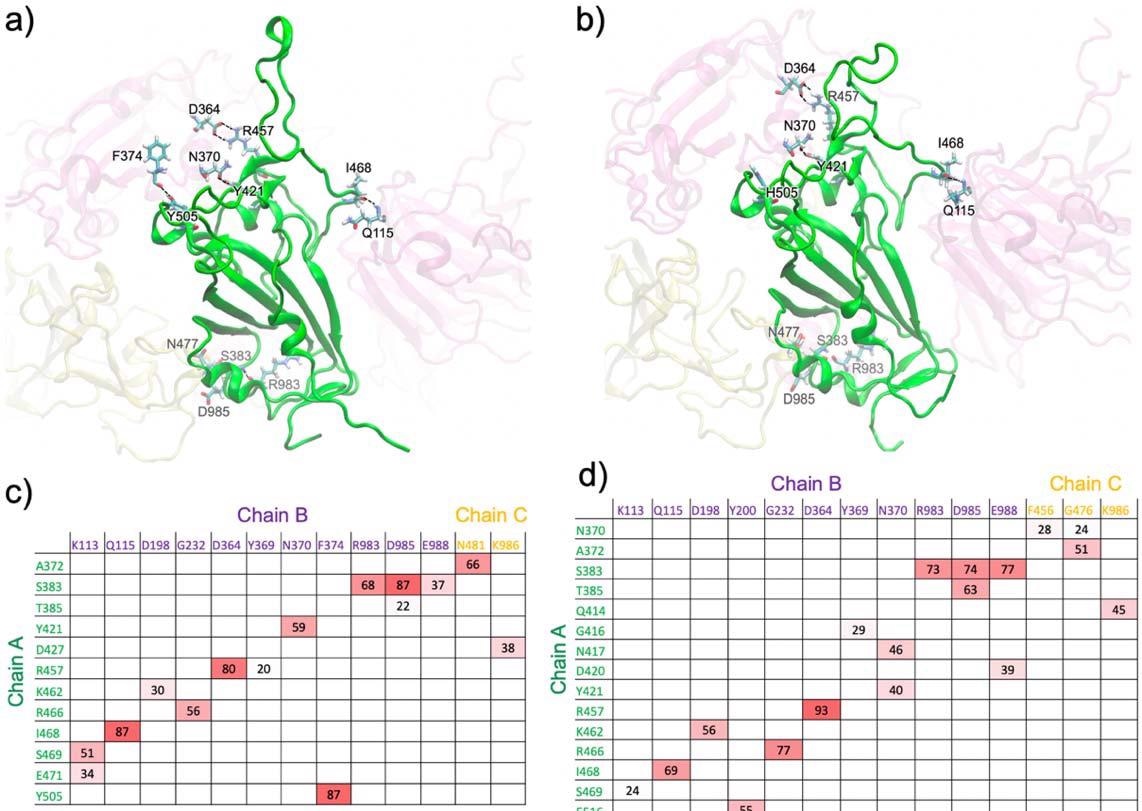
Major hydrogen bonds formed between the RBD of chain A (green) and the surrounding domains in the closed-form spike trimer for a) WT and b) Omicron. Additional interactions are shown in Fig. S1 (from different views). Hydrogen-bond pairs and % occupancies for the c) WT and d) Omicron, with the color scale from red (maximum) to white (minimum).
Impact of mutations on RBD structure and antibody binding
The structural analysis conducted in the study revealed significant alteration in omicron RBD structure compared to that in wild-type RBD. The primary structural change was observed in the motif that contains polar to hydrophobic mutations S371L, S373P, and S375F. These mutations significantly separated the residues 371 and 375 in the motif, which are potent antibody binding sites. This separation in the antibody binding sites can potentially reduce the efficiency of antibody binding, which in turn can facilitate the omicron variant to escape antibody-mediated neutralization. Further analysis revealed that almost all omicron RBD mutations are located at important epitopes, justifying omicron’s high immune escape ability.
The study identified three epitopes in the omicron RBD wherein mutations had caused significant induction in antigenicity compared to wild-type epitopes. The mutation-induced antigenic shifts observed in the omicron RBD can significantly reduce the sensitivity of wildtype-specific antibodies for these epitopes. However, these epitopes can induce a more robust immune response due to increased antigenicity.
Impact of mutations on ACE2 binding
The receptor-binding motif (RBM) of the omicron contains 10 mutations. To investigate the impact of these mutations on omicron RBD – ACE2 interaction, molecular dynamics simulations of the RBD – ACE2 complex were conducted in the study.
The findings revealed that the inter-protein hydrogen bond in the omicron is significantly higher than in the delta, indicating a more robust omicron RBD – ACE2 interaction. Four hydrogen bond pairs with more than 70% occupancy were identified in the omicron RBD – ACE2 complex. In contrast, only one such hydrogen bond pair was identified in the delta. These findings indicate that the interfacial interactions in the omicron are much stronger than that in the delta.
Compared to the wild-type virus, the omicron variant exhibited additional unique hydrogen bonds in the RBD – ACE2 interface. In contrast, no additional hydrogen bond was observed in the delta variant. Overall, these findings indicate a more efficient and stable omicron RBD – ACE2 interaction due to increased interfacial interactions.
Study significance
The study findings reveal that the omicron RBD has a higher binding affinity for ACE2 than the delta RBD. In addition, mutations present in the key omicron RBD epitopes may reduce the sensitivity of antibody binding, which in turn may increase the immune fitness of the variant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources