
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Several studies have demonstrated the role of humoral immunity in combating coronavirus disease 2019 (COVID-19) due to the ease of studying antibody molecules in vitro. Although T-cell receptor repertoire screening has provided insights into changes in T-cell responses elicited by COVID-19 vaccination or infection, studies which investigate effector functions of CTLs are sparse.
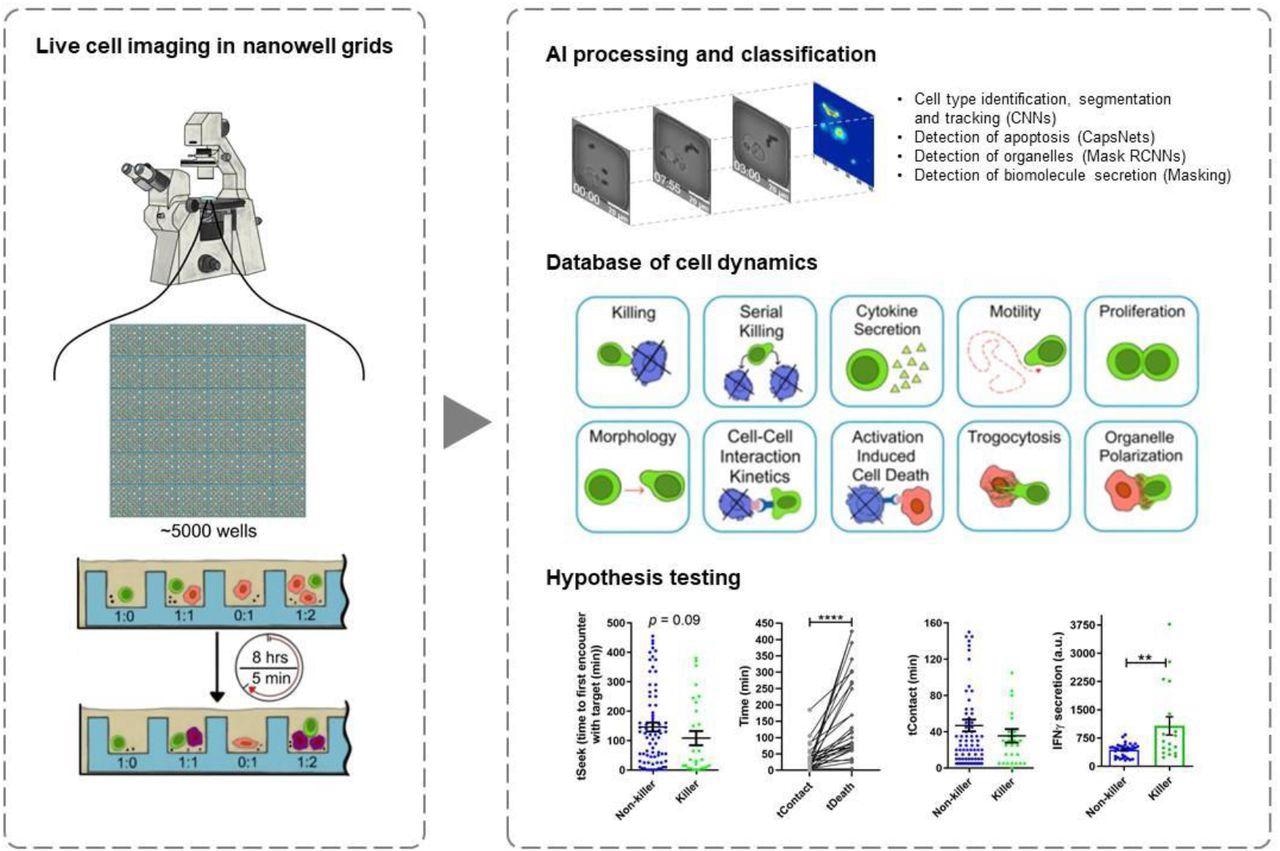
Mapping multiple functions of CTLs, including their activation, migration to infected cells, killer function, and secretion of cytokines, requires techniques that may examine the spatiotemporal changes in cellular behavior across thousands of cells.
Currently approved COVID-19 vaccines provide sterilizing immunity by inducing the generation of anti-SARS-CoV-2 neutralizing antibodies. However, vaccine-elicited antibody levels appear to wane over time, thereby putting vaccinated individuals at risk of acquiring COVID-19.
Improving the understanding of the cellular component of adaptive immunity mediated by CTLs in response to COVID-19 could help develop next-generation vaccines. These types of vaccines have the potential to provide broad and sustained cellular immunity against emerging SARS-CoV-2 variants.
About the study
In the present study, researchers used time-lapse imaging microscopy in nanowell grids (TIMING) based on artificial intelligence (AI) to integrate individual T-cell migration with effector functions analysis. The TIMING platform was also used to simultaneously study thousands of videos showing dynamic cell-cell interactions within nanowell grids, wherein SARS-CoV-2-specific CTLs eliminated multiple targets resembling SARS-CoV-2.
AI algorithms enabled high-throughput measurements of cell properties from single cells and cell-cell interactions that would otherwise be impossible to evaluate through conventional image acquisition. The TIMING platforms helped the researchers to visualize, monitor, and analyze the dynamic properties of SARS-CoV-2-specific T-cells in stimulator cells that were obtained from a convalescent individual and pulsed with spike (S) peptides to create targets that mimicked SARS-CoV-2 infection.
The researchers also quantified the CTL response to infections to distinguish T-cell behavior before, during, and after synapse formation. Some of the specific behavioral characteristics that were assessed include motility, morphology, and cytokine secretion until death occurred in the target cell.
The TIMING platform also identified unique groups of CTLs and distinguished various cell behaviors upon cell activation when adjusted to image sets of single T-cells at short intervals of five minutes.
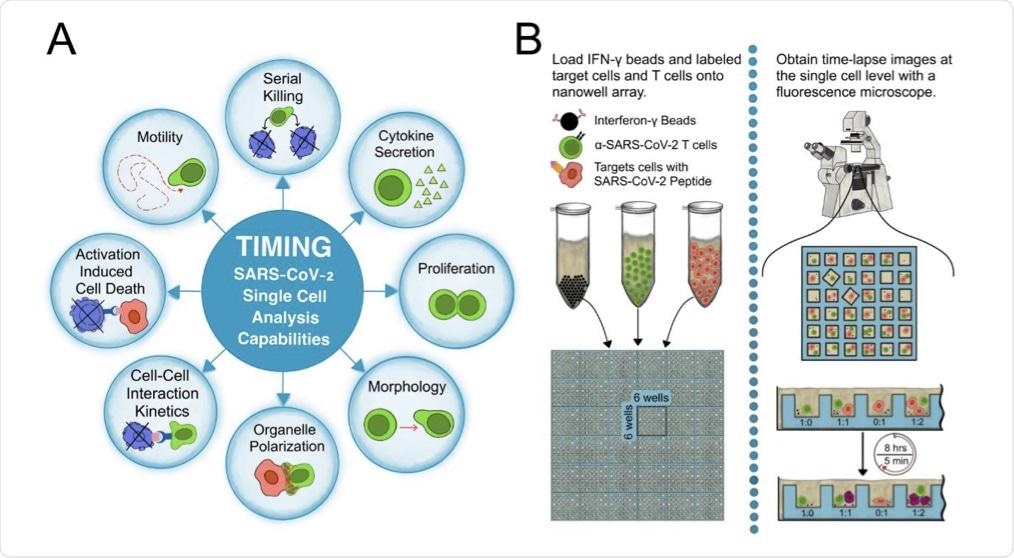
(A) Parameters measured by the TIMING platform. (B) The workflow of a TIMING assay.
Study findings
Upon tracking T-cells in nanowells, cells without conjugation to a target cell (1E:1T) showed increased motility. This observation indicates that T-cell motility was activated with co-incubation with target cells.
Within 270 nanowells bearing 1E:1T, 36% of the CTLs formed a stable synapse for 10 minutes or more. However, 27% of these CTLs induced apoptosis within the target cells, thereby demonstrating that there is an intra-clonal functional heterogeneity within CTLs and that a minority of effectors contribute to the majority of killing observed.
Analysis of nanowells bearing 1E:2T showed that 32% of T cells lysed one target, whereas 8% of CTLs participated in the serial killing and eliminated both targets.
Analyzing the frequency of activation-induced cell death (AICD) in nanowells with T-cell synapse formation (1E:1T) showed that killer CTLs had an elevated AICD frequency. This observation demonstrates that killer CTLs are more resistant to AICD than non-killer CTLs.
In addition to killing, CTLs are capable of secreting antiviral cytokines like interferon-gamma (IFNγ). Mapping the killing and cytokine secretion activities of individual T-cells that formed a synapse (1E:1T) showed that polyfunctional cells comprised only 11% of the total CTL population, whereas non-functional T-cells that neither killed nor secreted IFNγ were dominant.
When killer and non-killer T-cells were subjected to immunofluorescence to compare the rate of secretion of IFNγ, killer CTLs secreted significantly higher amounts of IFNγ. Taken together, these observations demonstrate that a subpopulation of T-cells is capable of both killing and IFNγ secretion.
Conclusions
To summarize, the researchers used AI-based imaging microscopy to show T-cell responses specific to SARS-CoV-. To this end, the researchers monitored the movement of thousands of T-cells and recognition of their target cells to reveal that T-cells are capable of serial killing and IFNγ secretion.
According to the authors, this is the first work that directly demonstrates that a subpopulation of CTLs specific for a conserved SARS-CoV-2 epitope is serial killers. This finding illustrates the power of T-cell mediated immunity and emphasizes the need to study cellular immune responses in the context of SARS-CoV-2 infection and vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Fathi, M., Charley, L., Cooper, L. J. N., et al. (2022). Cytotoxic T lymphocytes targeting a conserved SARS-CoV-2 spike epitope are efficient serial killers. bioRxiv. doi:10.1101/2022.01.24.477535. https://www.biorxiv.org/content/10.1101/2022.01.24.477535v1
- Peer reviewed and published scientific report.
Fathi, Mohsen, Lindsey Charley, Laurence JN Cooper, Navin Varadarajan, and Daniel D Meyer. 2022. “Cytotoxic T Lymphocytes Targeting a Conserved SARS-CoV-2 Spike Epitope Are Efficient Serial Killers.” BioTechniques 72 (4): 113–20. https://doi.org/10.2144/btn-2022-0016. https://www.future-science.com/doi/10.2144/btn-2022-0016.