The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), continues to cause immeasurable damage worldwide while efficient treatments remain scarce. The continued transmission of SARS-CoV-2 is largely due to the emergence of several mutant versions of the virus, such as the Alpha, Beta, Gamma, and Delta, and, more recently, Omicron variants.
Each of these five SARS-CoV-2 variants has been labeled as variants of concern (VOC) by the World Health Organization (WHO) as a result of their mutations’ ability to alter the binding, entry, and replication mechanisms of SARS-CoV-2. In fact, some of these mutations have increased the immune escape potential of SARS-CoV-2, which has led to a rising number of both re-infections and breakthrough infections.
Study: Differences between Omicron SARS-CoV-2 RBD and other variants in their ability to interact with cell receptors and monoclonal antibodies. Image Credit: Droneandy / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Both the SARS-CoV-2 Omicron variant, as well as its sub-lineage strain of BA.2, can evade vaccine-mediated responses and render them less effective. Similar immune evasion characteristics have been reported in plasma-based neutralization from convalescent individuals when challenged with these new SARS-CoV-2 strains.
Monoclonal antibodies (mAbs) offer a promising approach for the treatment of COVID-19; however, all SARS-CoV-2 variants require continuous bio-molecular and thermodynamic analysis of their interactions with available mAbs as a result of the evolution and mutation of the virus within hosts. To this end, studies evaluating the efficacy of current mAbs against these variants can assist in the development of new and specific mAbs, as well as their combinations, that target emerging SARS-CoV-2 variants.
In a recent study published on the preprint server bioRxiv*, researchers aimed to understand the relationship between the constantly evolving SARS-CoV-2 receptor-binding domain (RBD) and both human receptors and therapeutic targets. Special attention was given to the Omicron variant, as well as its sub-lineages BA.2, B.1.640.1, and B.1.640.2/IHU.
About the study
In the current study, the researchers used various molecular dynamics (MD) and Monte Carlo (MC) simulations in order to study various aspects of SARS-CoV-2 infection, including the biomolecular and thermodynamic parameters of virus-host interactions.
To this end, the researchers focused on two pairs of macromolecules in this regard. The first was the SARS-CoV-2 RBD from different variants with angiotensin-converting enzyme (ACE2) receptor polymorphisms like D355N, E37K, K26R, Y50F, D38V, G326E, Y83H, D509Y, H34R, K31R, E23K, H378R, K68E, E35K, I31K, N64K, and K26R+I31K.
In the second step, the researchers analyzed interactions between the RBD of variants and mAbs including LY-CoV555, LY-CoV016, LY-CoV1404, 4A8, SARS2- 38, AZD1061, AZD8895/COV2-2196, chAb25, S309, S230, REGN10933, REGN10987, CT-P59, EY6A, COR-101, Fab15033, B38, m336, m396, F26G19, CR3022, P2B-2F6, 80R, BD23, H11D4, and S2K146.
Study findings
All VOC RBDs had a higher affinity for the ACE2wt when compared to the SARS-CoV-2 wild-type RBD. As a result, this evolution improved the viral fitness of SARS-CoV-2 to bind to ACE2wt over time, with the Omicron RBD having the highest affinity for ACE2wt.
The RBDs of the B.1.640.1 and B.1.640.2/IHU sublineages of the Omicron variant behaved similarly to what was seen by the previous VOCs, thereby reducing concerns on the transmissibility of these new viral strains. When ranking the binding affinities from lowest to highest, the researchers characterized the variants into five distinct groups.
Whereas Group 1 included the wild-type, B.1.640.1, B.1.640.2, BA.2, mink, and Alpha strains, Group 2 included SARS-CoV-1, Beta, Epsilon, Gamma, and Mu strains. Group 3 included the Eta, Kappa, Iota, Mu-K417T, C1.2, and L452R+T478K strains of SARS-CoV-2, which was followed by the Delta and Omicron variants in Groups 4 and 5, respectively.
A plausible explanation for the increased transmissibility and rapid incubation of the Omicron variant was the increased RBDOmicron-ACE2 attraction. To this end, free energy calculations revealed that the Omicron variant had the strongest binding affinity as compared to any other SARS-CoV-2 variant, which was then followed by the Delta and Kappa variants. ACE2 polymorphisms like D509Y, G326E, H378R, K26R, and K68E increase the RBD-ACE2 binding affinities.
Among the mAbs included in this study, the binding affinity of S230 was highest for ACE2wt. The mAb S2K146 was seen to strongly bind competitively to the ACE2wt and potentially nullify viral infection through its interaction with the ACE2 receptor.
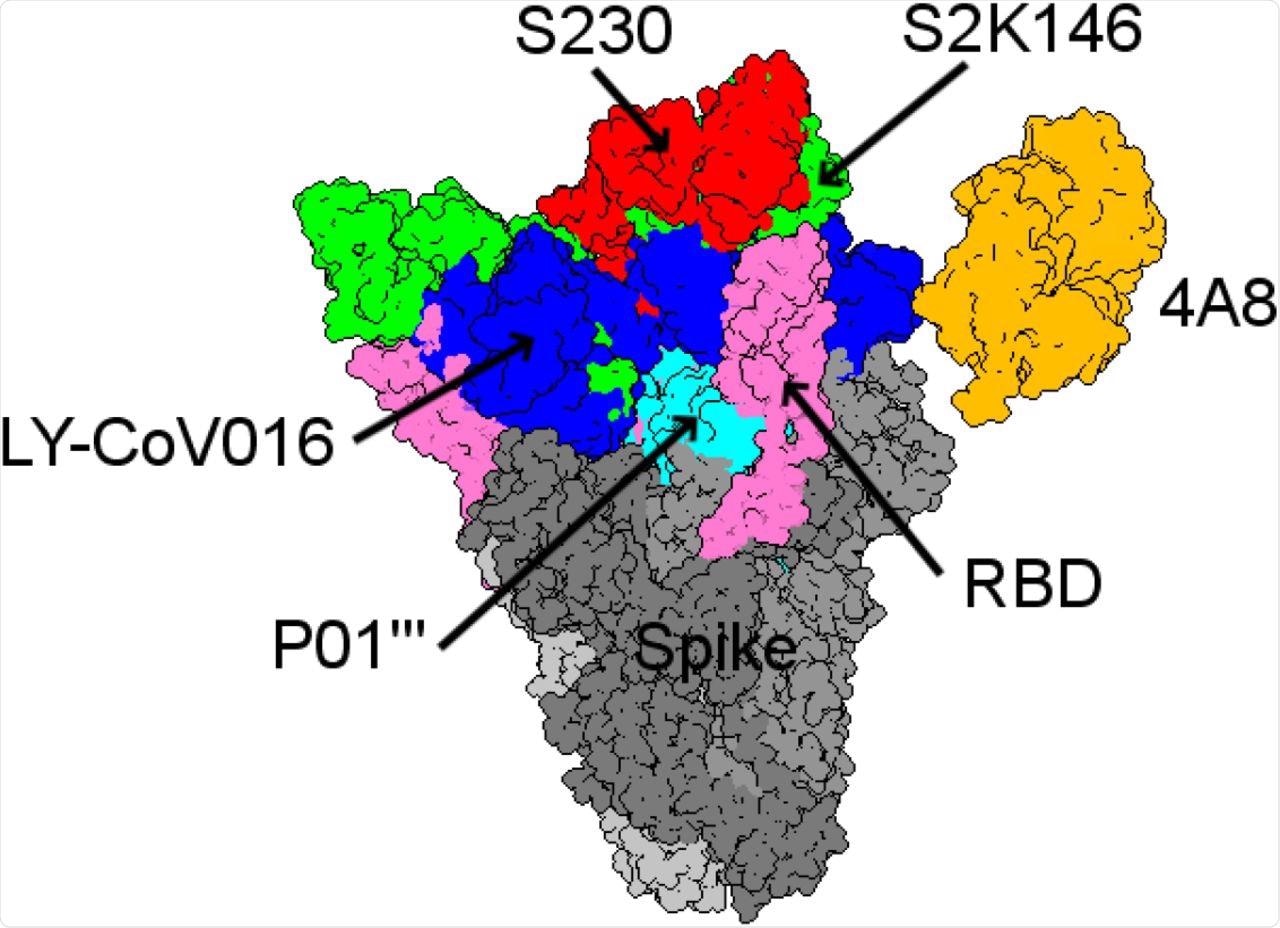
Structural comparison between the five best mAbs with the highest binding affinities from all the antibody landscapes.
Implications
The simulation models used in the current study showed that the SARS-CoV-2 Omicron variant had the strongest binding affinity to ACE2, which was followed by the Delta and Kappa variants. Furthermore, the researchers found that all tested mAbs in this study exhibited a strong binding capacity to the SARS-CoV-2 RBDs, including VOCs.
In particular, the researchers identified the S2K146 mAb to have a particularly promising future for both preventative and therapeutic purposes. Additional mAbs were also found to have similar binding affinities that could support their combined use with S2K146 in future studies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Giron, C. C., Laaksonen, A., & Barroso da Silva, F. L. (2022) Differences between Omicron SARS-CoV-2 RBD and other variants in their ability to interact with cell receptors and monoclonal antibodies. bioRxiv. doi:10.1101/2022.01.29.478316. https://www.biorxiv.org/content/10.1101/2022.01.29.478316v1.full.
- Peer reviewed and published scientific report.
Giron, Carolina Corrêa, Aatto Laaksonen, and Fernando Luís Barroso da Silva. 2022. “Differences between Omicron SARS-CoV-2 RBD and Other Variants in Their Ability to Interact with Cell Receptors and Monoclonal Antibodies.” Journal of Biomolecular Structure and Dynamics, July, 1–21. https://doi.org/10.1080/07391102.2022.2095305. https://www.tandfonline.com/doi/full/10.1080/07391102.2022.2095305.