SARS-CoV-2 contains a positive-sense, single-stranded (ss) RNA genome harboring two open reading frames, ORF1a and ORF1ab. Host cell ribosomes translate these ORFs to produce two polyproteins, and subsequently, 16 non-structural proteins (nsp1 - nsp16) form as a result of cleavage of the polyproteins. RNA-dependent RNA polymerase (RdRp) of nsp12, which forms the central element of replication-transcription complex (RTC), is the clinical target of various antiviral drugs used for coronavirus disease 2019 (COVID-19) treatment. Besides the RdRp domain, a Nidovirus RdRp-Associated Nucleotidyltransferase (NiRAN) domain is present at the N-terminus of nsp12.
Similar to eukaryotic messenger RNAs (mRNAs), the RNA genome of coronaviruses (CoVs) contains a 5′ guanosine cap at its first nucleotide. The 5′ cap is essential for stability, translation, and protection. The cap is methylated at 2′OH of the first nucleotide protecting the RNA from host immune responses, and therefore, the capping of RNA is critical to the viral life cycle.
The study
In the present study, the authors unraveled the mechanism of RNA capping in SARS-CoV-2. In eukaryotes, initially, a 5′-diphosphorylated RNA (5′-ppRNA) was produced by RNA triphosphatase (RTPase) removing γ-phosphate from the nascent 5′-pppRNA. This diphosphorylated RNA was capped with a GMP transferred from GTP by guanylyltransferase (GTase) to form the core (GpppN-RNA).
The core was methylated at N7 by a guanine-N7-methyltransferase (N7-MTase) and finally by a nucleoside-2′-O-methyltransferase (2′-O-MTase) at 2′OH position of ribose on the first RNA nucleotide. According to studies in CoVs, RTPase, N7-MTase, and 2′-O-MTase activities are exhibited by nsp13, nsp14, and nsp16, respectively. Based on these reports, it has been speculated that RNA capping in CoVs is similar to the eukaryotic mechanism but remains uncharacterized.
Findings
The NiRAN domain was implicated in a kinase-like process called NMPylation. The researchers noted the NiRAN-dependent NMPylation of nsp9 confirmed with nuclear magnetic resonance (NMR) spectroscopy. Mutating Asn1 with Ala or Asp decreased the NMPylation of nsp9. The authors then constructed a 10-mer RNA (5′-pppRNALS10) with the first 10 bases of SARS-CoV-2 leader sequence (LS10) and incubated it with nsp12 and nsp9. They observed that a monophorylated RNALS10 (5′-pRNALS10) was incorporated into nsp9, and similarly, the mutation of Asn1 reduced the RNA incorporation into nsp9. Further, substituting the first nucleotide with a different nucleotide too diminished RNAylation.
The nsp9-5′-pRNALS10 intermediate was purified and incubated with GDP and nsp12. Treatment with GDP caused deRNAylation of the intermediate, transferring the 5′-pRNALS10 to GDP, creating GpppA-RNALS10, and the reaction was conditional to the presence of a functional NiRAN domain. The authors reported the methylation of the cap by the transfer of methyl group from the donor, S-adenosyl methionine (SAM). The nsp 14 and nsp16 of SARS-CoV-2 exhibited N7-MTase and 2′-O-MTase activities, respectively, and additionally, nsp16 requires nsp10 for its activity. The authors incubated the NiRAN-synthesized cap with SAM in the presence of nsp14 and nsp16/10 complex. Treatment with nsp14 produced 7MeGpppA-RNALS10, and subsequent incubation of 7MeGpppA-RNALS10 with nsp10/16 complex resulted in 7MeGpppA2′-O-Me-RNA.
Using cryo-electron microscopy, the authors provided structural insights into RNA capping by the NiRAN domain. nsp9's first four residues extended into the NiRAN domain's active pocket and formed electrostatic and hydrophobic interactions. Then Asn1 of nsp9 accepts 5′-pppRNA onto its N-terminus. The C-terminus and Leu4 of nsp9 formed hydrophobic connections with β-sheet of NiRAN domain. nsp9's N-terminus was involved in electrostatic interactions with the NiRAN domain's negatively charged pocket on the surface. The researchers constructed mutated nsp9 (mutations: N1A, N1D, and N2A) and nsp12 (K73A, D218A, and D760A), infected cells, and subsequently infected cells; the viral quantification showed a 400 to 4000-fold reduction in viral load.
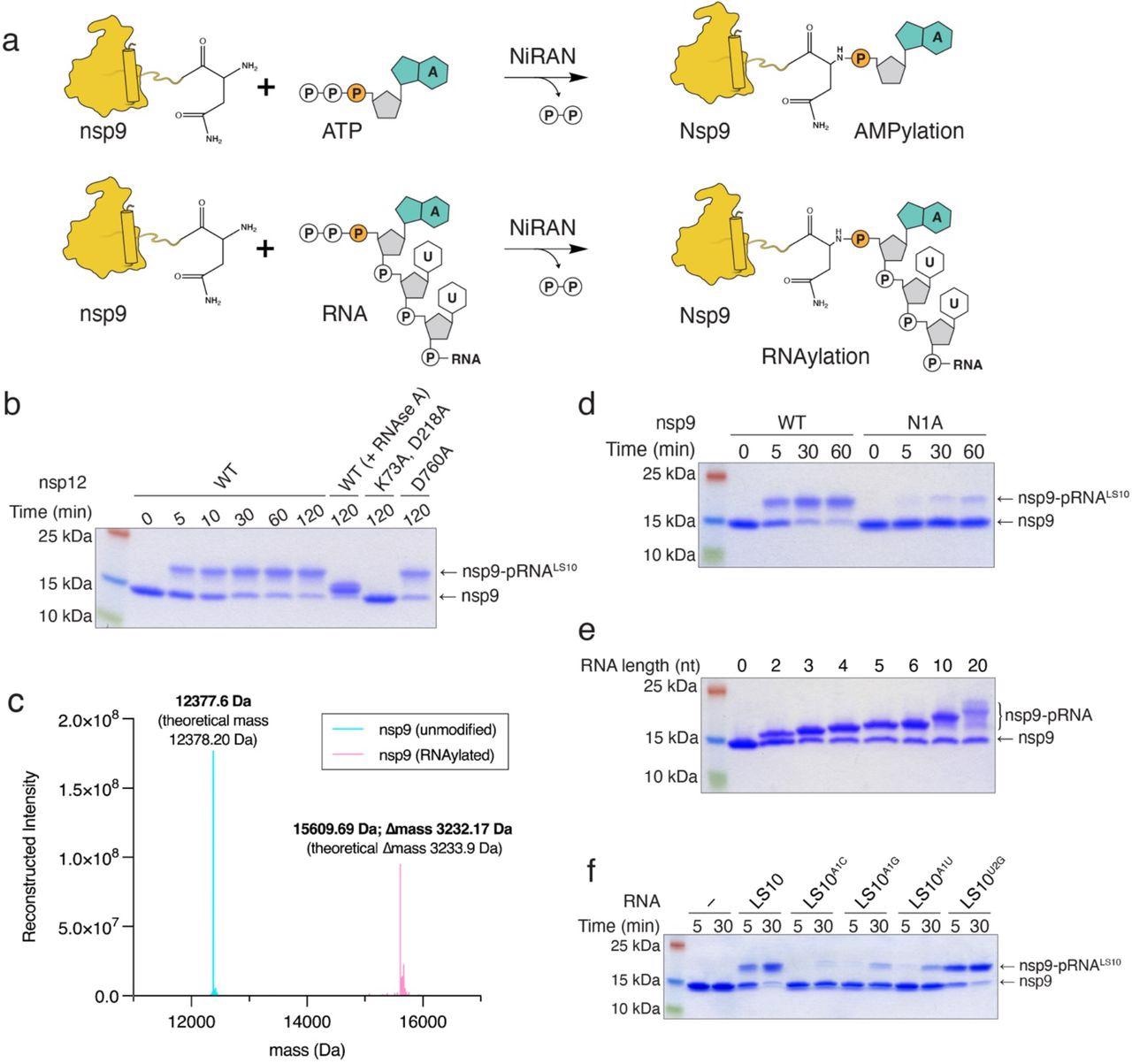
The NiRAN domain RNAylates nsp9. a. Schematic depicting the nsp9 AMPylation reaction (top) and the proposed nsp9 RNAylation reaction (bottom). b. Time-dependent incorporation of RNA into nsp9 by WT nsp12, the NiRAN mutant (K73A, D218A), or the polymerase mutant (D760A). Reaction products were analyzed by SDS-PAGE and Coomassie staining. Samples were also treated with RNAse A. c. Intact mass LC/MS spectra (overlayed) of unmodified nsp9 (cyan) or nsp9 after incubation with 5’-pppRNALS10 and WT nsp12 (pink). The theoretical and observed masses are shown in the insets. The △mass of 3233.17 Da corresponds to monophosphorylated RNALS10 (5’-pRNALS10). d. Time-dependent incorporation of 5’-pRNALS10 into nsp9 or the nsp9 N1A mutant. Reaction products were analyzed as in b. e. Incorporation of different lengths of 5’-pppRNAs into nsp9 by the NiRAN domain. Reaction products were analyzed as in b. f. Time-dependent incorporation of RNAs with substitutions in the first and second base into nsp9 by the NiRAN domain. Reaction products were analyzed as in b. Results shown are representative of at least 2 independent experiments.
Conclusions
The proposed RNA capping mechanism is as follows. Initially, the nascent RNA (5′-pppRNA) binds in NiRAN domain′s active site, and subsequently, the monophosphorylated 5′-pRNA is transferred and linked to nsp9 through phosphoramidite bond. The GTase activity of nsp13 synthesizes GDP from GTP, and the GDP deRNAylates nsp9-5′pRNA regenerating the nsp9, forming the GpppA-RNA cap.
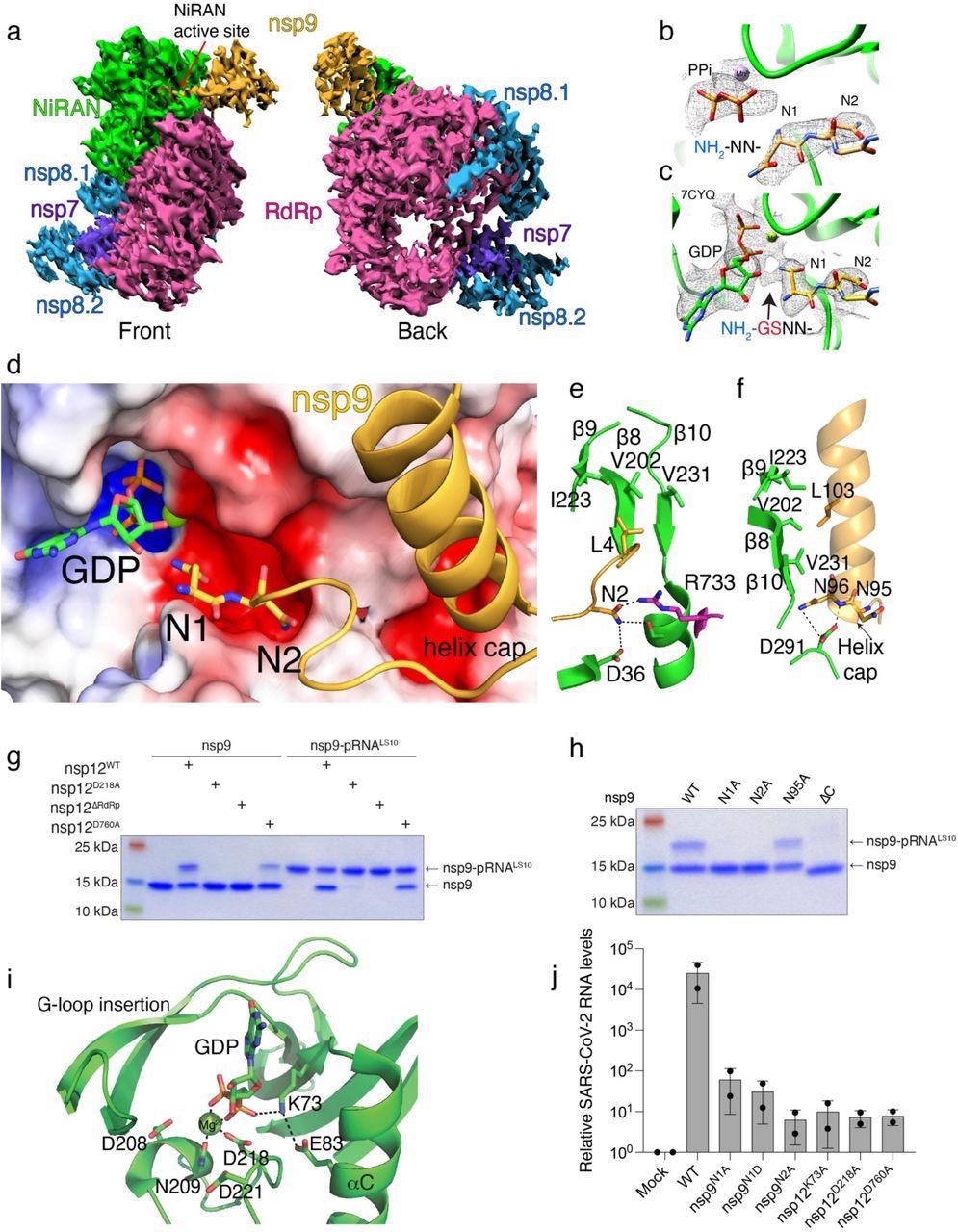
Structural and genetic insights into RNA capping by the kinase domain. a. Front and back views of nsp12/7/8/9 cryo-EM maps, with respect to the NiRAN domain. The NiRAN domain is in green, the RdRp in magenta, nsp7 in violet, nsp8 in light blue and nsp9 in gold. b, c. Coulomb density maps of the N-terminus of nsp9 from this study (b) and by Yan et al. (c) 21 (PDB ID:7CYQ). The NiRAN domain is shown in green and nsp9 is in gold. The arrow in (c) indicates additional density that likely corresponds to unmodeled Gly-Ser residues at the non-native N-terminus of nsp9. d. Electrostatic surface view of the NiRAN active site from 7CYQ bound to nsp9 (gold). The N-terminus and the C-terminal helix of nsp9 are shown. The electrostatic surface of nsp12 is contoured at 5 kT. e. Cartoon representation depicting the interactions between the nsp9 N-terminus (gold) with the β8-β9-β10 sheet in the NiRAN domain (green). Asn2 in nsp9 forms electrostatic interactions with Asp36 in the NiRAN domain and Arg733 in the RdRp domain (magenta). PDB ID 7CYQ was used. f. Cartoon representation depicting the interactions between the nsp9 C-terminal helix (gold) and the β8-β9-β10 sheet in the NiRAN domain (green). Interactions between Asn95/96 in nsp9 and D291 in the NiRAN domain are indicated. PDB ID 7CYQ was used. g. Incorporation of 5’-pRNALS10 into nsp9 and deRNAylation of nsp9-pRNALS10 by WT nsp12, the NiRAN mutant (D218A), the polymerase mutant (D760A), or the isolated NiRAN domain (residues 1-326; △RdRP). Reaction products were analyzed as in Fig. 2b. h. Incorporation of 5’-pRNALS10 into nsp9 (or the indicated mutants) by nsp12. Reaction products were analyzed as in Fig. 2b. i. Cartoon representation of the NiRAN active site. Catalytic residues and GDP are shown as sticks, Mg2+ is a green sphere, and interactions are denoted by dashed lines. j. Relative viral yields from WT or mutant SARS-CoV-2 viruses bearing indicated mutations in nsp9 and nsp12. Data represent averages of two biological replicates. Error bars, SD. Results shown in g and h are representative of at least 2 independent experiments.
Finally, this cap is methylated by successive methylations by N7-MTase and 2′-O-MTase to produce the final functional product, 7MeGpppA2′-O-Me-RNA. Moreover, the functions of nsp13 are proposed to use ATP hydrolysis energy for helicase activity to unwind double-stranded (ds) RNA GTP hydrolysis to release GDP, catalyzing the cap formation. The NMPylation reaction of nsp9 was reversible in the presence of inorganic pyrophosphate, but RNAylation was irreversible, indicating that RNAylation was a physiologically relevant reaction during the viral capping process.
The researchers conclude, "In summary, we have defined the mechanism by which SARS-CoV-2 caps its genome and have reconstituted this reaction in vitro using non-structural proteins encoded by SARS-CoV-2. Our results uncover new targets for the development of antivirals to treat COVID-19 and highlight the catalytic adaptability of the kinase domain."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
The mechanism of RNA capping by SARS-CoV-2, Gina J Park, Adam Osinski, Genaro Hernandez, Jennifer Eitson, Abir Majumdar, Marco Tonelli, Katherine Henzler-Wildman, Krzysztof Pawlowski, Zhe Chen, Yang Li, John Schoggins, Vincent S. Tagliabracci, bioRxiv 2022.02.07.479471, DOI: 10.1101/2022.02.07.479471, https://www.biorxiv.org/content/10.1101/2022.02.07.479471v1
- Peer reviewed and published scientific report.
Park, Gina J., Adam Osinski, Genaro Hernandez, Jennifer L. Eitson, Abir Majumdar, Marco Tonelli, Katie Henzler-Wildman, et al. 2022. “The Mechanism of RNA Capping by SARS-CoV-2.” Nature, August, 1–8. https://doi.org/10.1038/s41586-022-05185-z. https://www.nature.com/articles/s41586-022-05185-z.