Since the discovery of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), several viral variants have emerged, some of which have exhibited superior virulence and transmissibility as compared to the original strain.
The World Health Organization (WHO) has classified these more virulent strains of SARS-CoV-2 as variants of concern (VOC) or variants of interest (VOI) in accordance with their infectiousness, mutations, virulence, and capacity to evade immune responses generated by immunization or natural infection.

Study: Genomic and Virological Characterization of SARS-CoV-2 Variants in a Subset of Unvaccinated and Vaccinated U.S. Military Personnel. Image Credit: Emily C. McCormick / Shutterstock.com
Background
To date, five SARS-CoV-2 VOCs have been identified, which include the B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and B.1.1.529 (Omicron) strains. Recently, the Omicron variant has become the dominant circulating strain in most countries around the world.
Prior to the emergence of the Omicron variant, the scientists characterized the Delta variant to be more transmissible than the Alpha variant. To this end, studies indicated that the Delta variant caused increased hospitalization rates among unvaccinated individuals.
The emergence of SARS-CoV-2 VOCs has also increased reports of vaccine breakthrough infections (VBIs). Although real-time reverse-transcription polymerase chain reaction (qRT-PCR) has proved to be extremely efficient in detecting COVID-19, one of the limitations of this assay is that it detects the viral ribonucleic acid rather than the virus itself. This is problematic, as viral loads estimated by qRT-PCR do not always correlate with the amount of viable virus in a given sample.
Within a laboratory setting, scientists detect infectious viruses through in vitro viral culture on susceptible cell lines. Although this method is effective and provides insights related to viral shedding, it is labor-intensive and requires biosafety level 3 (BSL-3) facilities when handling SARS-CoV-2.
Generally, Vero cells and their subclones, such as are Vero E6 cells which express human transmembrane serine protease 2 (TMPRSS2), are commonly used to culture SARS-CoV-2. Plaque or median tissue culture infectious dose (TCID50) assays are used to quantify viable viruses.
Although several studies have described methods to measure the amount of viable virus shed by individuals infected with various SARS-CoV-2 variants, more research is required to characterize virus levels among vaccinated and unvaccinated individuals. This is essential, as different studies have followed different methods and have focused on varied samples from different geographic locations.
About the study
In a new Frontiers in Medicine study, researchers discuss the genomic and virological characterization of SARS-CoV-2 variants among unvaccinated and vaccinated U.S military personnel. The authors evaluated nasal swab samples that were collected between March 2020 and early November 2021 from a cohort of U.S. military personnel and beneficiaries stationed worldwide as a part of the Defense Health Agency's (DHA) Global Emerging Infections Surveillance (GEIS) program.
SARS-CoV-2 positive samples were determined and subsequently subjected to next-generation sequencing. The subsets were analyzed to estimate viable viruses.
Study findings
A significant fraction of the VBI cases in the current study were associated with the Delta variant. The researchers observed that the increased transmissibility of this variant was partly due to higher amounts of virus shedding.
Interestingly, among VBIs, the authors reported a 50-fold increase in the viable virus in the nasal swab samples from Delta variant cases as compared to cases involving other variants. Similarly, a 40-fold increase in the viable virus was found in nasal swab samples from VBIs involving Delta as compared to unvaccinated personnel infected with other variants.
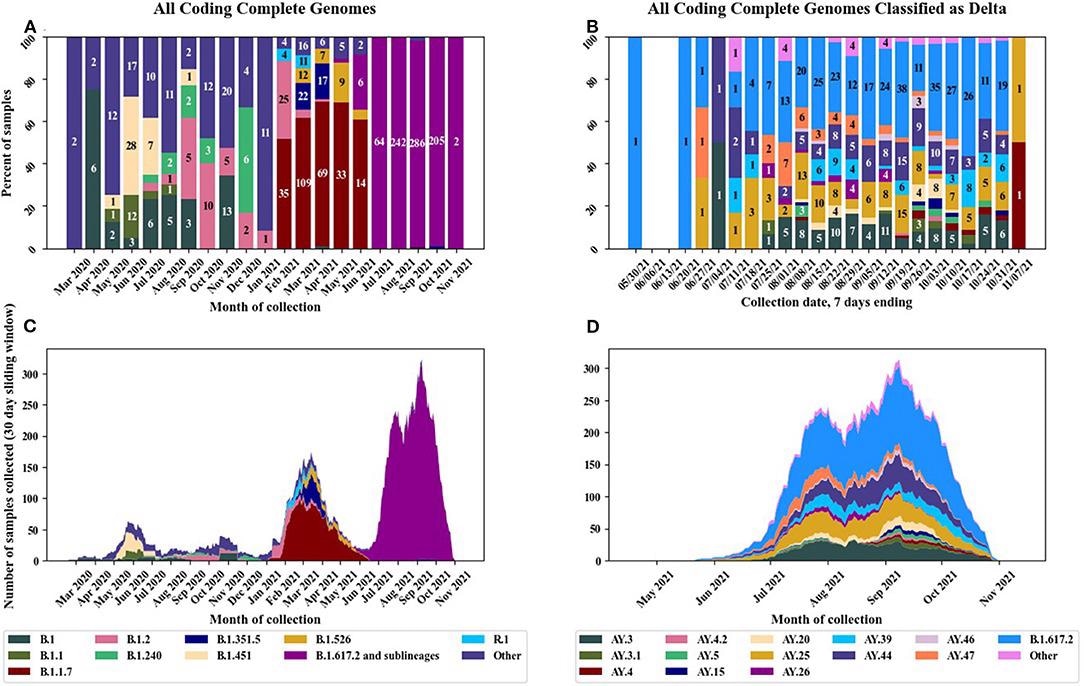
Pango lineages of circulating SARS-CoV-2 strains in the U.S. military before and after implementation of the EUA vaccines. (A) Distribution of Pango lineages by month for all samples, (B) distribution of Delta variant sublineages by month, (C) stacked chart displaying all lineages per month, and (D) stacked chart displaying Delta variant sublineages by month.
Individuals remained unvaccinated because, at that time, no vaccines had received approval from global regulatory bodies. The findings from this study are in line with previous studies that included the rest of the population in the U.S., where the Delta variant was the dominantly circulating strain.
Scientists reported that the Delta variant contains the R203M mutation, whereas the Omicron variant contains the R203K mutation, which might be the reason for the increased fitness of these variants. Prior research has indicated that samples from Delta variant infected individuals showed increased viral RNA levels and a longer period of viral shedding as compared to samples obtained from people infected with other variants.
The researchers also observed similar levels of viral RNA from vaccinated and unvaccinated individuals. In the future, more studies are required to determine the duration of viral shedding and viral burden associated with the Omicron variant.
The scientists analyzed their results across vaccine manufacturers and detected significantly higher levels of viable virus in samples from individuals who received the Pfizer and Johnson & Johnson vaccines. These individuals were found to experience more VBIs due to Delta variant infection than unvaccinated personnel with non-Delta infections.
Conclusions
One of the limitations of this study was the unavailability of metadata for the clinical samples. This hindered proper interpretation of the results with regard to differences in age, pre-existing conditions, and days from exposure or symptom onset.
Further, the study cohort included young military personnel between the ages of 18 and 40 who are required to maintain proper physical conditions for their duty. Therefore, a lack of diversity could also lead to biased results.
The current study has emphasized the importance of characterizing the SARS-CoV-2 genome and viable viral titers of all emerging SARS-CoV-2 variants isolated from unvaccinated and vaccinated personnel.
Journal reference:
- Smith, R. D., Singh, C., Green, J., et al. (2022) Genomic and Virological Characterization of SARS-CoV-2 Variants in a Subset of Unvaccinated and Vaccinated U.S. Military Personnel. Frontiers in Medicine. doi:10.3389/fmed.2021.836658.