The evolution of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causal agent of the coronavirus disease 2019 (COVID-19) pandemic, has occurred due to genomic mutations. As a result, several SARS-CoV-2 variants have emerged, which have been classified as variants of concern (VOC) and variants of interest (VOI) in accordance with the virulence and transmissibility of the new variant with respect to the original SARS-CoV-2 strain.
According to a recent report, the SARS-CoV-2 Omicron (B.1.1.529) variant has been characterized as 2.7- to 3.7-times more transmissible than the Delta (B.1.617.2) pre-Delta variants.
Study: SARS-CoV-2 variants show a gradual declining pathogenicity and pro-inflammatory cytokine stimulation and an increasing antigenic and anti-inflammatory cytokine induction. Image Credit: Viacheslav Lopatin / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Scientists have reported that the SARS-CoV-2 infection rate is significantly high in individuals between the ages of 18 and 49 years. In addition, the SARS-CoV-2 Omicron variant has been reported to be highly transmissible, even among fully vaccinated adults.
However, the mortality rate is significantly low in individuals infected with the Omicron variant compared to other SARS-CoV-2 variants. To date, the exact cause of the high transmission rate and reduced severity of the Omicron variant is not well documented.
Some studies have indicated that the presence of many mutations in the SARS-CoV-2 spike (S) protein region of the Omicron variant has led to a high rate of transmission and decreased disease severity and death. Most studies have focused on the interactions between the receptor-binding domain (RBD) of the S protein and human angiotensin-converting enzyme 2 (hACE2) to understand the increased transmissibility of the Omicron variant.
These studies have revealed that the Omicron RBD strongly binds to hACE2, while others indicated low affinity of the Omicron RBD to hACE2. Thus, more studies are needed to understand the strength of the Omicron RBD-hACE2 interaction and the stability of this complex.
About the study
A new study posted to the bioRxiv* preprint server focuses on understanding the factors associated with the ability of the Omicron variant to infect a wide range of age groups. Herein, scientists used various bioinformatics tools to analyze randomly selected genomic sequences of different SARS-CoV-2 strains, including the Gamma, Delta, and Omicron variants.
The authors also analyzed why SARS-CoV-2 pre-Omicron variants did not infect or cause severe symptoms in young adults. To this end, the scientists revealed that there are many possible immunological reasons for the differences mentioned above in disease manifestation between the Omicron and pre-Omicron variants.
In this study, the researchers analyzed several SARS-CoV-2 structural proteins, including the S, membrane (M), nucleocapsid (N), and envelope (E) proteins, as well as the pathogenicity-associated with accessory proteins ORF3a, ORF7a, ORF7b, ORF8, and ORF10)of the Omicron variant. This provided a better understanding of the role of these proteins in higher transmissibility and decreased disease severity of the Omicron variant compared to other SARS-CoV-2 VOCs.
Study findings
Based on a sequence-based analysis, the researchers reported that RBD, M, N, ORF3a, and ORF7a are the main proteins associated with the antigenic variation. Concerning the pathogenic property, the gradual decline of SARS-CoV-2 pathogenicity in strains began with the original strain and was followed by the Gamma, Delta, and Omicron variants.
Interestingly, the scientists revealed that compared to the Delta variant, the Omicron variant exhibited 20% lower genomic pathogenicity and 32% lower pathogenicity at structural protein levels. This finding is in line with a recent study, which reported that the Omicron variant displays significant antigenic variation as compared to other variants.
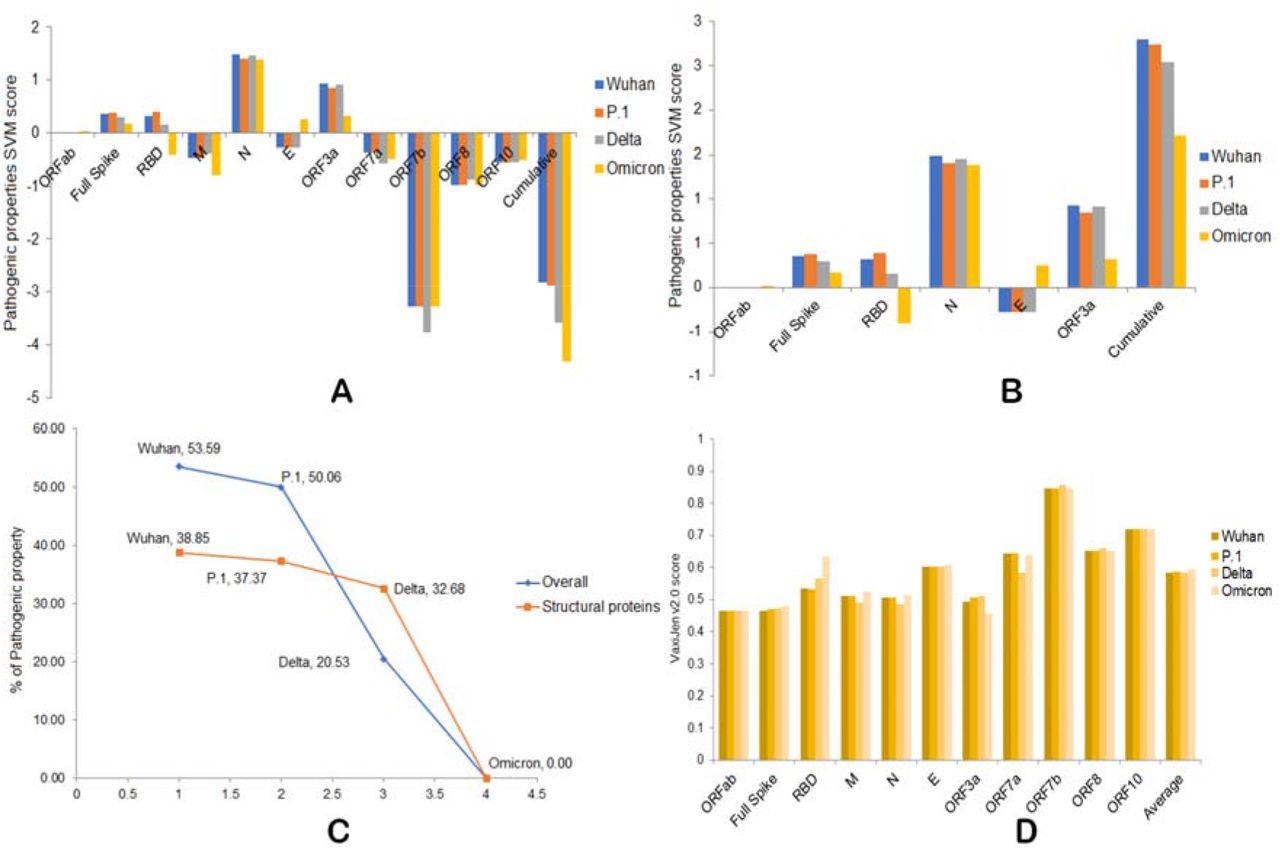
A) Overall pathogenic properties of the four variants. B) Overall pathogenic properties of structural proteins from the four variants. C) The percentage of decreased pathogenicity of the four variants as compared to Omicron. D) Antigenic properties of four variants.
Moreover, the scientists revealed that the Omicron variant could be the best strain of SARS-CoV-2 for developing new COVID-19 vaccines based on the attenuated virus. Additionally, the S RBD of the Omicron variant could be a potential candidate to design viral vector-, peptide-, and nucleic acid-based COVID-19 vaccines.
The S protein may also produce a reduced level of pro-inflammatory and interleukin-6 (IL-6) inducing epitopes compared to the Delta and other SARS-CoV-2 variants. The N protein also plays a vital role in producing these peptides.
The N and ORF3a proteins were also found to play an essential role in interferon γ (IFN-γ) and IL-4 induction differences in these variants. Increased production of anti-inflammatory IFN-γ and IL-4 induction capacity of the Omicron variant as compared to other SARS-CoV-2 variants was also reported. Specifically, the order of production was Omicron = Gamma > Wuhan > Delta and Omicron ≥ Delta > P.1Gamma > Wuhan.
A robust interaction was also found to occur between the Omicron N protein and human DDX21.
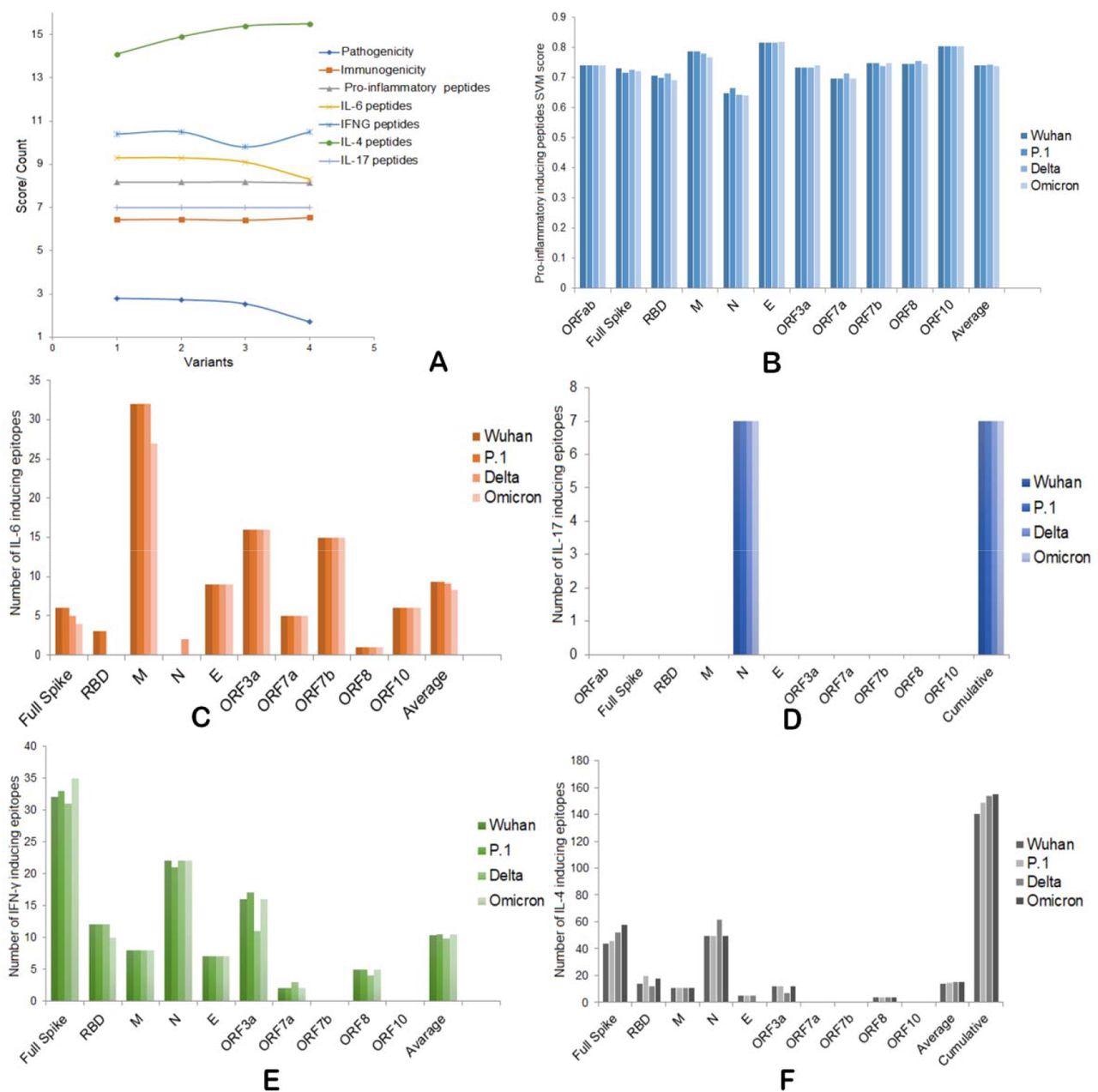
A) Overall pathogenic, immunogenic, IFNs, and ILs induction abilities of four SARS-CoV-2 variants. B) Pro-inflammatory epitope production scores of four variants. C) IL-6 inducing epitope counts of four variants. D) IL-17 inducing epitope counts of four variants. E) Number of IFN-γ inducing epitopes by four variants. F) Number of IL-4 inducing epitopes by four variants.
Conclusions
The findings from the current study suggest that the SARS-CoV-2 Omicron variant manifests milder infection with less severity due to its increased ability to elicit IFN-γ and IL-4, along with a decreased ability to induce pro-inflammatory cytokines and IL-6 as compared to other SARS-CoV-2 variants.
Additionally, a strong RBD-hACE2 interaction could be the reason for the hyper-transmissibility of the Omicron variant. The scientists also revealed that attenuated replication of the Omicron variant is responsible for the reduced disease severity and death rates.
Notably, the E protein is strongly associated with the replication attenuation of this variant. As the E protein is unstable, it may decrease replication or maturation of new viral Omicron Taken together, the bioinformatics strategy used in this analysis could be implemented to study the dynamics of other viruses as well.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Barh, D., Tiwari, S., Gomes, L. G. R., et al. (2022) SARS-CoV-2 variants show a gradual declining pathogenicity and pro-inflammatory cytokine stimulation and an increasing antigenic and anti-inflammatory cytokine induction. bioRxiv. doi:10.1101/2022.02.15.480592. https://www.biorxiv.org/content/10.1101/2022.02.15.480592v2.
- Peer reviewed and published scientific report.
Barh, Debmalya, Sandeep Tiwari, Lucas Gabriel Rodrigues Gomes, Cecília Horta Ramalho Pinto, Bruno Silva Andrade, Shaban Ahmad, Alaa A. A. Aljabali, et al. 2022. “SARS-CoV-2 Variants Show a Gradual Declining Pathogenicity and Pro-Inflammatory Cytokine Stimulation, an Increasing Antigenic and Anti-Inflammatory Cytokine Induction, and Rising Structural Protein Instability: A Minimal Number Genome-Based Approach.” Inflammation, October. https://doi.org/10.1007/s10753-022-01734-w. https://link.springer.com/article/10.1007/s10753-022-01734-w.