A new study under consideration at a Nature Portfolio Journal and published on the preprint server Research Square* discusses the emergence of new variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that can escape antibody-mediated neutralization, as opposed to other Sarbecoviruses. The researchers of this study conclude that these mutations likely arose under selection pressure due to antibodies elicited by infection, vaccination, or both, in the human population on a large scale.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic that began in December 2019 quickly reached every country of the world with devastating mortality and morbidity. The causative pathogen SARS-CoV-2 is a sarbecovirus, which is similar to the earlier SARS-CoV that triggered an outbreak almost two decades ago.
SARS-related coronaviruses are known to be harbored by bats and possibly pangolins. As compared to these viruses, SARS-CoV-2 mutated into several variants that have been identified in quick succession since late 2020, some of which have been denoted as variants of concern (VOCs). SARS-CoV-2 VOCs are more transmissible and/or able to evade neutralization by antibodies elicited by the vaccines, by prior infection, or both.
To date, five SARS-CoV-2 VOCs have been identified, of which include the Alpha, Beta, Gamma, Delta, and Omicron variants. The Omicron variant has the most mutations in the spike protein as compared to any other SARS-CoV-2 variant, with 32 and 15 mutations identified in its spike and receptor-binding domain (RBD) of the spike, respectively. The SARS-CoV-2 Omicron variant can also escape neutralizing antibodies to a much higher extent than previous VOCs, which has contributed to the enhanced transmissibility of the virus.
Direct comparisons of neutralization capacity between Omicron, the ancestral strain, and other VOCs are possible through the use of conventional live virus neutralization test (cVNT) or spike pseudovirus VNT (pVNT). Despite their utility, these tests are cumbersome and labor-intensive, with multiple assays available for each test that has hindered the ability to make accurate comparisons.
To overcome these issues, the current study used a multiplex surrogate virus neutralization test (sVNT) platform to examine the antibody-escape capacity of all known SARS-CoV-2 VOCs, as well as of other bat and pangolin Sarbecoviruses. This test measures neutralizing antibodies (nAbs) to the RBD, which accounts for over 90% of their total neutralizing activity.
Study findings
The researchers coupled the use of this platform, which can handle up to 16 sVNTs simultaneously, with 20 panels of human serum. This included five and eleven SARS-CoV-2 clade 1 and 2 RBD variants, respectively.
Clade 1 included SARS-CoV and bat viruses like Rs2018B, LYRa11, RsSHC014, and WIV1, while clade 2 included the ancestral SARS-CoV-2 strain, the five SARS-CoV-2 VOCs, several SARS-CoV-2 few variants of interest (VOI), along with the bat RaTG13 and pangolin GX-P5L.
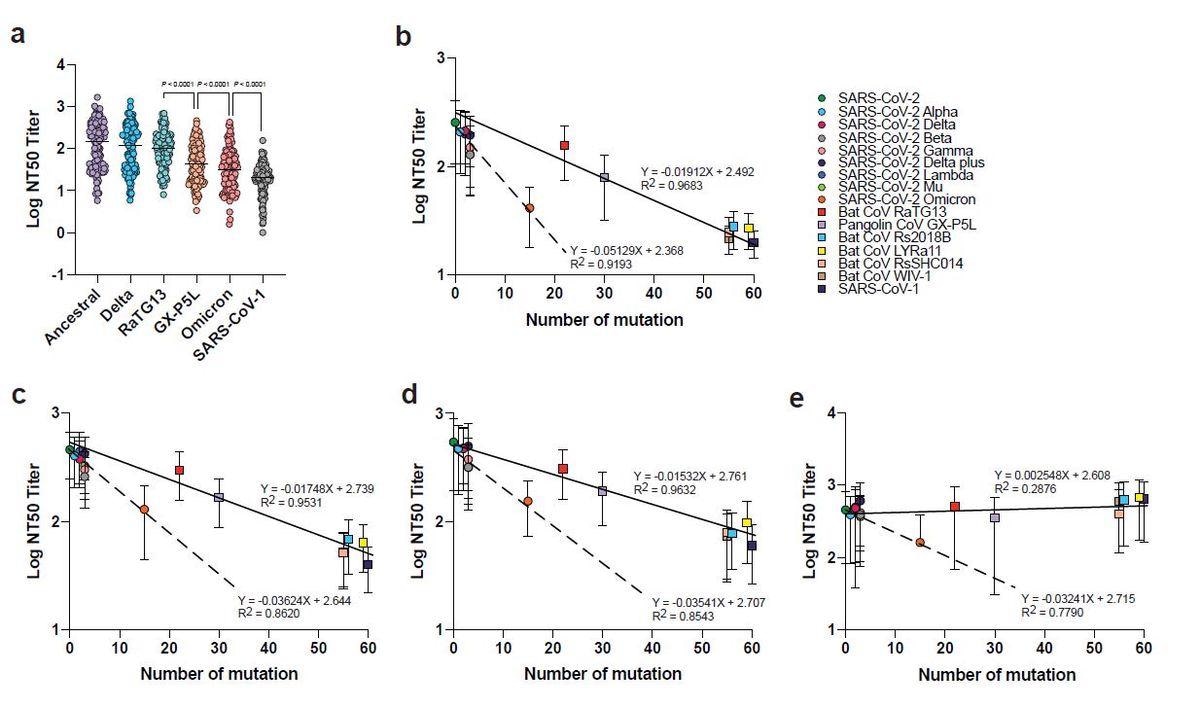
Neutralization escape in relation to the number of RBD mutations. a, Neutralization titers derived from multiplex sVNT (n = 124). The effect of RBD mutations on NAb escape for different serum panels including those with b, two doses of mRNA vaccines, c, three doses of mRNA vaccine, d, vaccinated individuals with breakthrough infection, and e, BTN162b2-vaccinated SARS survivors. Paired two-tailed student’s t-tests were used in a. Line in a indicates median. Linear regression analysis in b-e was performed using GraphPad prism. Line and dotted line in b-e indicate linear regression plot on SARS-CoV-2 with pre-emergent sarbecoviruses and VOCs, respectively.
The most closely related sequences were observed in the SARS-CoV-2 Alpha, Delta, Beta, and Gamma variants, in that order, with the Lambda interspersed between the first two and the last two. The Delta, Mu, Omicron, RaTG13, and GX-P5L variants, as well as the other bat/pangolin coronaviruses subsequently followed in order of their similarity. SARS-CoV was found to be the most distantly related virus.
Interestingly, the SARS-CoV-2 Omicron variant is farthest in sequence identity from SARS-CoV due to its 65 RBD mutations. The Omicron RBD also showed the weakest binding to the human angiotensin-converting enzyme 2 (ACE2) receptor on the host cell.
Convalescent sera from patients with prior infection with ancestral and Beta variants showed a greater than 10-fold and about seven-fold reduced neutralization, respectively, of Omicron compared to prior Omicron infection. A smaller reduction was observed with prior Delta infection.
With vaccinated patients who had received two doses of the Pfizer or Moderna COVID-19 vaccines, the level of nAbs against Omicron was reduced by more than eight-fold and 5.7-fold, respectively. Other vaccines, including the inactivated viral vaccines and viral vector-based vaccines, induced a lower neutralizing as response compared to the messenger ribonucleic acid (mRNA) vaccines.
With a third booster dose, the two mRNA vaccines and the adenoviral vector vaccine from AstraZeneca showed a marked rise in nAb titers to all SARS-CoV-2 variants, but not inactivated virus vaccines. Even with this rise, nAb titers to Omicron were still lower when compared to the ancestral variant by four- to five-fold depending on the exact regimen of either three (homologous or heterologous) mRNA vaccine doses or two viral vector vaccine doses with an mRNA/heterologous booster.
In people with hybrid immunity, as defined as those with a history of prior SARS-CoV-2 infection and were vaccinated, nAb reduction to Omicron was on the order of two- to five-fold. The greatest reduction was following vaccination and Delta breakthrough infection, whereas the lowest reduction in nAb titers was with Omicron breakthrough infections.
Both Omicron and Delta breakthrough infections induced similar nAb titers to Omicron; however, no samples had neutralizing activity against clade 1 sarbecovirus. SARS-CoV-vaccinated individuals had no neutralizing activity against clade 1 showed that pan-Sarbecovirus nAbs occur only with SARS-CoV vaccination and not natural infection with SARS-CoV-2. Interestingly, these samples also showed some loss of neutralizing activity against Omicron.
Antigenically, the Omicron RBD is more distant from SARS-CoV-2 than the bat CoV RaTG13 and pangolin CoV GX-P5L, even though it has 93% sequence identity with 15 mutations as compared to 90% and 86.6% sequence similarity, respectively, with 22 and 30 mutations, respectively. Omicron also escapes nAbs more efficiently than these two viruses.
SARS-CoV-2 VOCs escaped nAbs more significantly than the pre-emergent Sarbecoviruses, thus demonstrating the need for more RBD mutations in the latter to achieve the same immune escape potential. In fact, Omicron shows an unexpectedly high nAb escape relative to the number of mutations in the RBD when compared to SARS-CoV survivors who had received the Pfizer vaccine.
With the use of hyperimmune rabbit sera, Omicron showed a nine-fold reduction in neutralization. The sera raised against bat and pangolin RaTG13 and GX-P5L had the highest nAb titers; however, these titers were about seven-fold and four-fold reduced against Omicron, respectively.
Implications
Despite extensive vaccine coverage in the developed world, as well as the widespread of SARS-CoV-2 infection globally, the Omicron variant has shown the ability to escape nAbs. This has made it necessary to provide an mRNA booster to reach the Omicron-protective level of nAbs, irrespective of the vaccine used earlier. However, the AZ viral vector vaccine could also provide a booster if given after two doses of inactivated viral vaccines.
The overall neutralizing titers were higher after breakthrough infection against all variants; however, Delta was superior to Omicron in this respect.
Besides demonstrating the power of this platform, the current study demonstrates that the Omicron variant is exceptionally competent to escape host immunity, even more than GX-P5L with twice as many RBD mutations. However, it could not induce nAbs against clade 2 sarbecoviruses.
Only individuals who had prior SARS-CoV infection and received the SARS-CoV-2 mRNA vaccines were found to have pan-sarbecovirus nAbs, though even these individuals had lower neutralizing titers against Omicron.
If newer vaccines are developed, they must act independently of the RBD mutations. Otherwise, variants like Delta and Omicron may continue to emerge under selection pressure from antibodies developed against the ancestral RBD. In fact, developing vaccines that immunize across SARS-CoV-2 clades may be the best way to induce pan-sarbecovirus immunity.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.