The coronavirus disease 2019 (COVID-19) pandemic has caused an unprecedented health emergency for the past two years resulting in more than 447 million infections and over 6 million deaths to date. SARS-CoV-2, the etiologic agent of COVID-19, has mutated throughout the pandemic, with new and more infectious variants constantly emerging. As a result, the World Health Organization (WHO) has classified these variants into variants of concern (VOC) or interest (VOI), or under monitoring (VUM).
 Study: Identifying SARS-CoV-2 Variants of Concern through saliva-based RT-qPCR by targeting recurrent mutation sites. Image Credit: NIAID
Study: Identifying SARS-CoV-2 Variants of Concern through saliva-based RT-qPCR by targeting recurrent mutation sites. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The mutation rate of SARS-CoV-2 is estimated to be 1 x 10-3 substitutions per base per year under neutral genetic drift conditions. Although most substitutions are insignificant, some confer selective advantages for (enhanced) transmission and immune evasion. Whole-genome sequencing (WGS) can differentiate SARS-CoV-2 variants with exceptional resolution. Although WGS is considered the gold standard for variant differentiation, it is not feasible for real-time application of large-scale surveillance as it is expensive and time-consuming. These limitations necessitate a cost-effective method for the surveillance of SARS-CoV-2 variants at a population level.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) has been implemented for public surveillance of SARS-CoV-2 variants such as the Alpha, Beta, Gamma, and Delta VOCs. As the Alpha VOC was detected based on S-gene target failure (SGTF), researchers exploited target gene failure for detecting single nucleotide polymorphisms (SNPs) or deletions among VOCs. Interestingly, using competitive probes for mutation and reference sequences in RT-qPCR assay increases its specificity.
The study
The present study evaluated TaqPath (spike SNP) assays for saliva samples for the K417T, E484Q, E484K, and L452R mutations. RT-qPCR assays for ORF1aΔ3675-3677 and SΔ69-70 were developed in-house. As the latest Omicron variant carries the SΔ69-70 and L452R mutations, these assays were used to identify Omicron’s emergence in December 2021. The cross-validation of the assays was performed using WGS results.
Synthetic ribonucleic acids (RNAs) of six SARS-CoV-2 strains (B.1, B.1.1.7, B.1.351, P.1, B.1.617.1, and B.1.617.2) were serially diluted 10-fold for estimating analytical sensitivity of the RT-qPCR assays. Similarly, analytical specificity was assessed by carrying out SΔ69-70 and ORF1aΔ3675-3677 deletion assays and spike SNP assays for K417T, E484K, E484Q, and L452R on the synthetic RNAs from six strains.
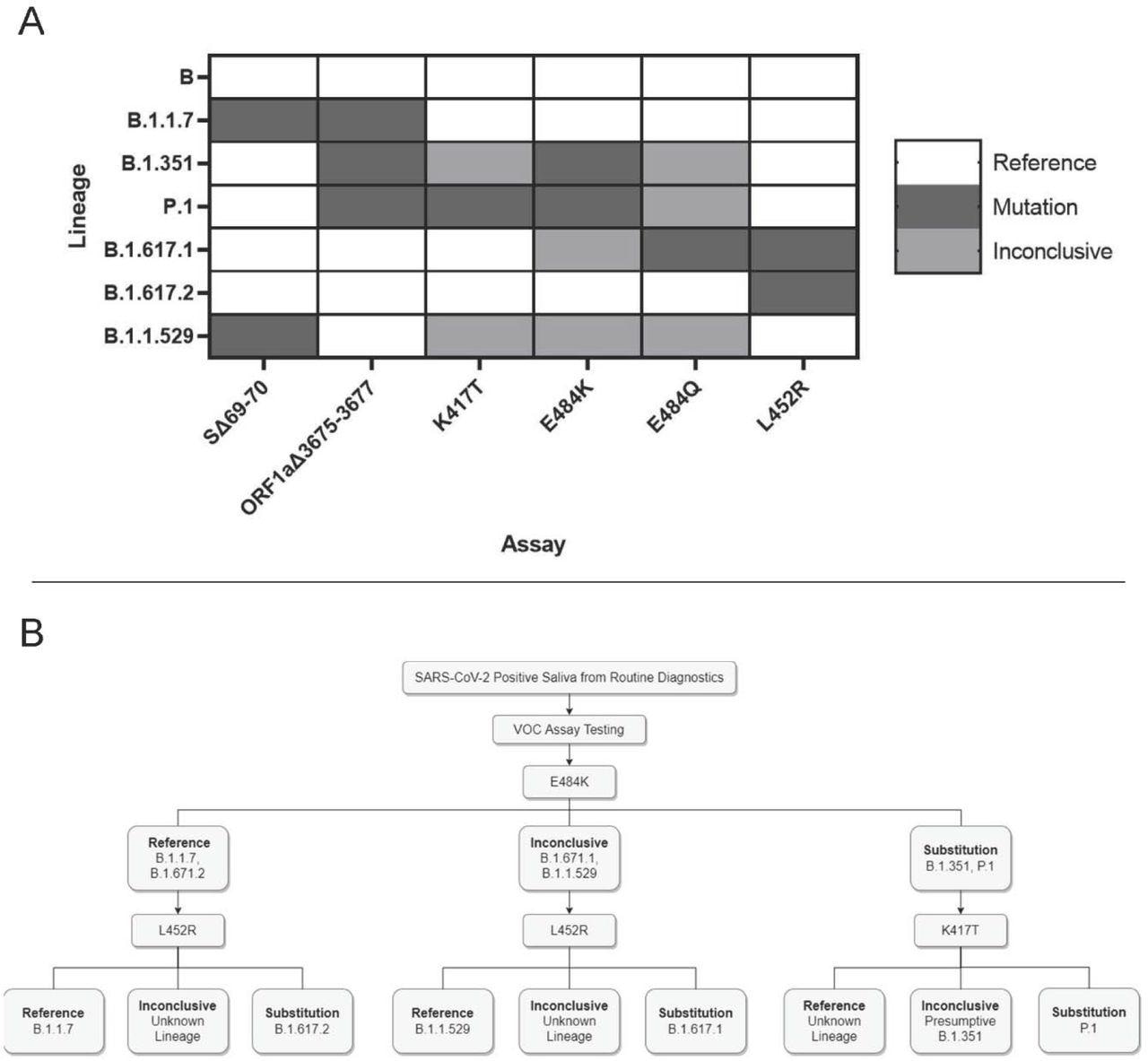
Application and interpretation of differential VOC assays. 3A. VOC strain typing by mutation site. Each strain will produce a different combination of results from the six assays. Strains with an alternate allele at the mutation site will produce inconclusive results. 3B. Example strain typing workflow using minimal steps. Saliva samples that are determined positive by routine diagnostic testing are analyzed by various assays that produce differential results for each VOC.
Findings
The authors estimated the efficiency of mutation probes to be in the range of 89.5 – 112.04%. The detection limit (LoD) for the SΔ69-70 deletion assay was 40 genome copies per assay, while it was four genome copies per assay for ORF1aΔ3675-3677, E484K, K417T, L452R, and E484Q, including the control gene (nucleocapsid [N] gene).
No amplification failure or cross-reactivity was detected for any deletion assay. The fluorescent output varied with a low output for the ORF1aΔ3675-3677 assay. Although SNP assays showed no cross-reactivity, amplification failure, as expected, for strains without both mutation and reference sequences. These results indicated the high specificity of the SNP assays to synthetic RNAs.
The deletion and spike SNP assay results were compared with WGS results to ascertain their clinical sensitivity and specificity. Total accuracy, individual probe accuracy, and positive (PPV) and negative predictive values (NPV) were also estimated for the deletion and spike SNP assays.
SΔ69-70 assay yielded a total accuracy of 93.6%, while it was 68% for the ORF1aΔ3675-3677 assay. Clinical sensitivities were 94.82% (SΔ69-70 assay) and 95.65% (ORF1aΔ3675-3677 assay), and the clinical specificity and PPV were 100% for both the deletion assays. Total accuracy was higher than 96.4% for all spike SNP assays; clinical sensitivity and NPV were 100% for all assays except L452R. Also, clinical specificity ranged between 95.05 – 99.44% for SNP assays.
Additionally, 162 SARS-CoV-2-positive samples were collected between December 7 – 16, 2021. Of that, 13 samples were identified with the L452R assay for reference sequence at this locus. Next, the SΔ69-70 assay performed on these 13 specimens identified deletions in 11 of them. WGS confirmed that these 13 sequences were of the Omicron variant. Similarly, 183 additional samples were screened between December 17 – 22, 2021, and the authors found 107 prospective Omicron-positive samples.
Conclusions
The current study evaluated the efficacy of RT-qPCR assays for six mutation sites of SARS-CoV-2 variants on saliva samples. The authors designed two deletion assays for SΔ69- 70 and ORF1aΔ3675-3677 sites and validated four TaqPath Spike SNP assays.
The SNP assays produced accurate results even in samples with low viral load suggesting that saliva samples do not confound the assays’ fidelity. However, the current strategy is insufficient for comprehensive strain typing, and increasing the number of mutation sites could maximize strain identification. To this end, the authors are working on validating SNP assays for the N501Y, G339D, and K417N mutations.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Identifying SARS-CoV-2 Variants of Concern through saliva-based RT-qPCR by targeting recurrent mutation sites, Rachel E Ham, Austin R Smothers, Rui Che, Keegan J Sell, Congyue Annie Peng, Delphine Dean, medRxiv 2022.03.02.22271785; DOI: https://doi.org/10.1101/2022.03.02.22271785, https://www.medrxiv.org/content/10.1101/2022.03.02.22271785v1
- Peer reviewed and published scientific report.
Ham, Rachel E., Austin R. Smothers, Rui Che, Keegan J. Sell, Congyue Annie Peng, and Delphine Dean. 2022. “Identifying SARS-CoV-2 Variants of Concern through Saliva-Based RT-QPCR by Targeting Recurrent Mutation Sites.” Edited by Heba H. Mostafa. Microbiology Spectrum 10 (3). https://doi.org/10.1128/spectrum.00797-22. https://journals.asm.org/doi/10.1128/spectrum.00797-22.