Notwithstanding many successes of the global vaccination campaign against COVID-19, at the moment, there is a lack of effective drugs to treat individuals infected with SARS-CoV-2, which means the identification of promising compounds is currently a research priority.
We know that SARS-CoV-2 has a relatively large, positive-sense, and single-stranded genome made of ribonucleic acid (RNA), consisting of 12 functional open reading frames (ORF) responsible for encoding four structural proteins, six accessory proteins, and 16 non-structural proteins.
These non-structural proteins are basically the machinery for viral replication, including RNA-dependent RNA polymerase, proteases, primase, helicase, nucleases, and a plethora of enzymes involved in RNA capping. One of the more salient ones is a bifunctional protein known as NSP14.
A specific role of NSP14 in SARS-CoV-2
Of note, SARS-CoV-2 NSP14 contains an N-terminal DEDDh-type exoribonuclease (ExoN) domain and a C-terminal SAM-dependent guanine-N7 methyltransferase (N7-MTase) domain. This is important to emphasize, as the aforementioned ExoN plays a role in RNA proofreading, which aids in preserving the integrity of the viral genome by removing mismatched nucleotides.
This activity is substantially enhanced by the interaction of NSP14 with the activator protein and zinc-binding protein known as NSP10. Although this interaction greatly enhances the nuclease, but not the methyltransferase activity, both enzymatic activities appear to be pivotal for the viral life cycle and, therefore, can be targeted with antiviral therapeutics.
The complexes of these two proteins have been solved by crystallography and cryogenic electron microscopy, and more recently, there are studies taking a deep dive into the relationship of SARS-CoV-2 NSP14-ExoN domain in complex with NSP 10.
In this new study, Dr. Nergis Imprachim, Dr. Yuliana Yosaatmadja and Dr. Joseph A Newman have initially elucidated the crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 to 1.7-angstrom resolution and then compared it with the structure of NSP14/NSP10 complexes.
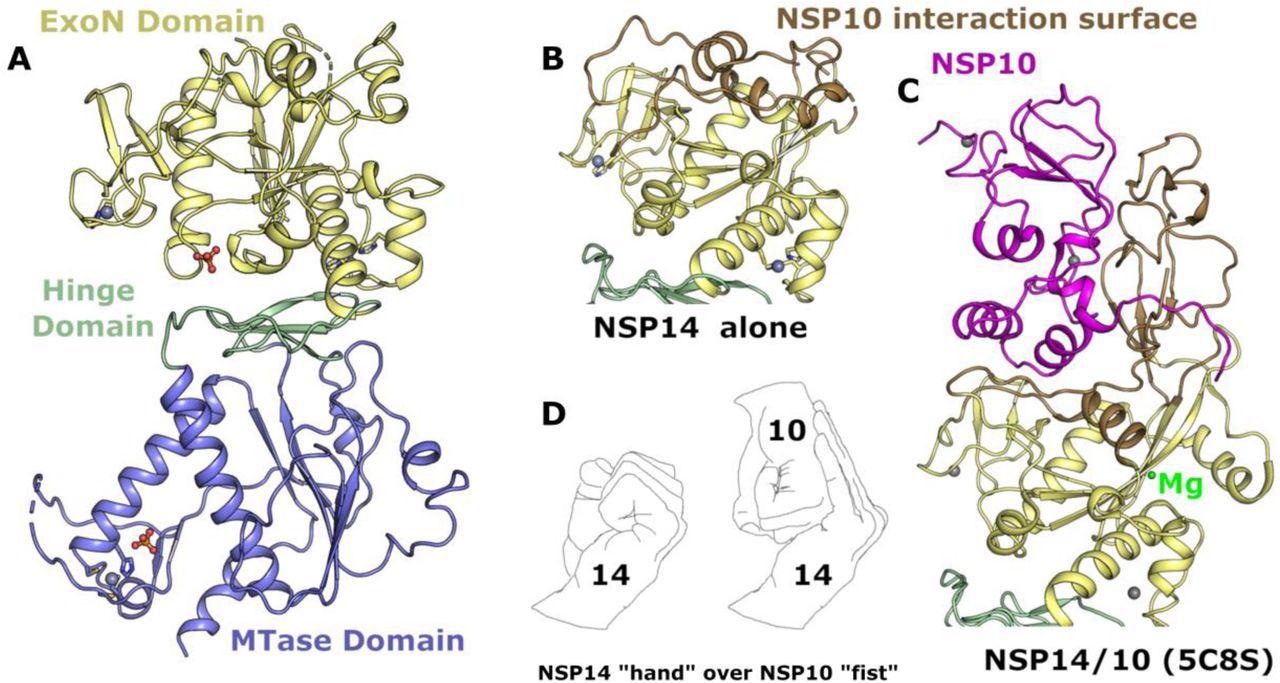
Structure of NSP14 in the absence of NSP10. (A) Overall NSP14 structure with the ExoN domain colored yellow, hinge domain in green and MTase domain blue, zinc ions are shown as grey spheres. (B) Close-up view of the ExoN domain with regions that were observed to shift conformation highlighted in “sand” color. (C) Structure of the ExoN domain of SARS-CoV NSP14 in complex with NSP10 (shown in pink) viewed from the same orientation as for panel B. (D) The interaction between NSP14 and NSP10 has been described as “hand over fist”, by the same logic the “fingers” of NSP14 collapse back toward the core resembling a closed “fist”.
Identifying conformational changes
The most significant output from this study was the identification of conformational changes that are observed within the NSP14 ExoN domain upon binding of NSP10. This includes substantial movements and helix to coil transitions that facilitate the formation of the ExoN active site and at the same time, provide an explanation of the nuclease activity stimulation by NSP10.
Furthermore, various conformational changes were also seen in the MTase active site within a specific interacting loop that has a crucial role in viral mRNA capping. New insights into MTase enzymatic activity have also been observed, opening the door for further work in this field.
In addition, the researchers have used high-resolution crystals to perform X-ray fragment screening of NSP14, disclosing 72 hits bound to potential sites of inhibition of the ExoN and MTase domains. As one of the most conserved targets in the coronaviral genome, NSP14 may be an excellent target for broad-spectrum antiviral agents.
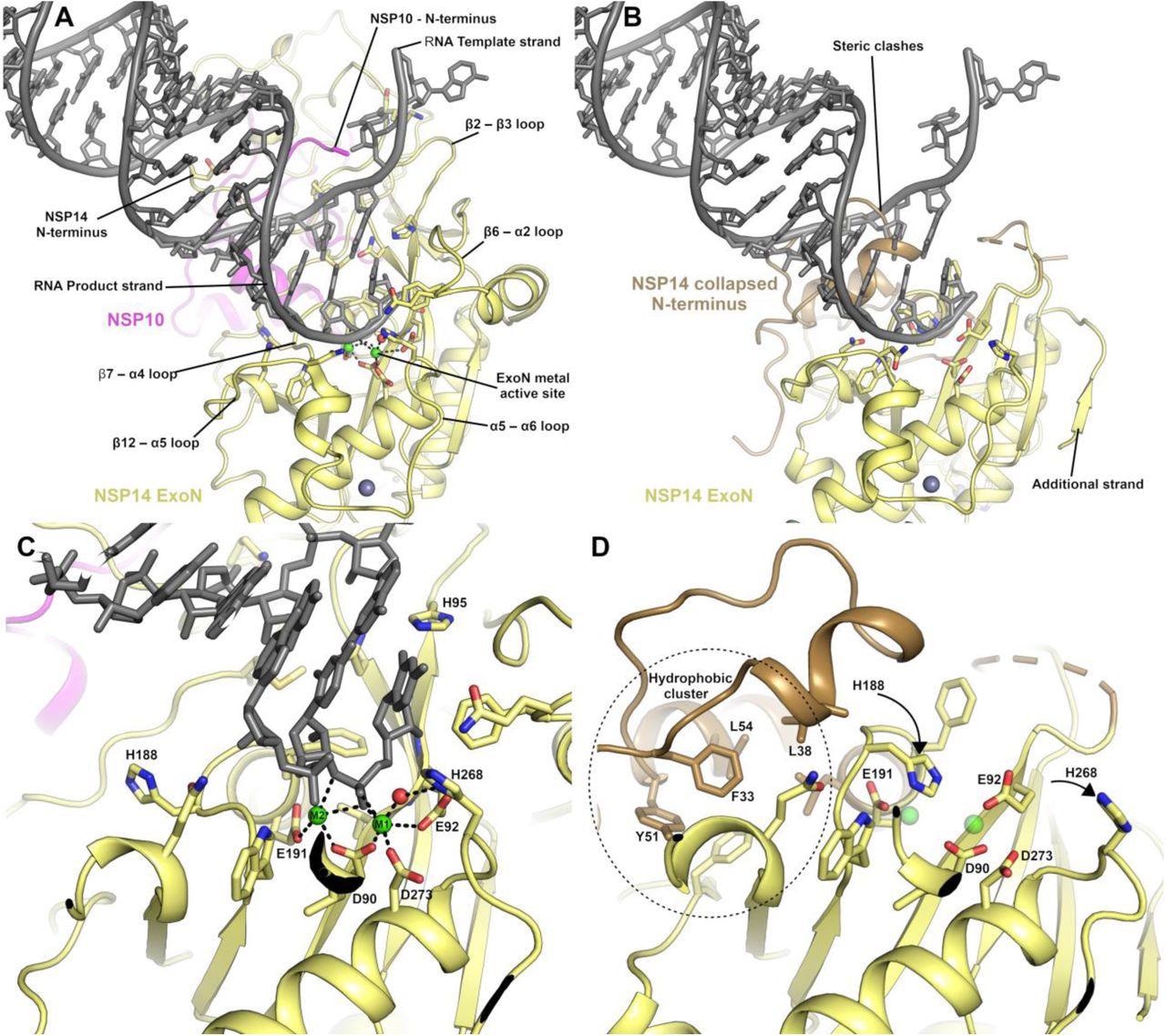
The NSP14 ExoN active site is formed upon binding to NSP10. (A) View of the ExoN domain of the NSP14 NSP10 complex bound to a mismatch containing double-stranded RNA molecule as recently solved by cryo EM9. Key residues in the catalytic center are shown in the stick format and the secondary structure elements that form the wider active site are labeled. (B) View of the NSP14 ExoN domain in the absence of NSP10, the mismatch containing RNA is shown for reference and forms significant steric clashes with residues that constitute the collapsed N-terminus (shown in sand color). (C) Zoomed-in view of the active site catalytic center of the NSP14 NSP10 RNA complex with metal ion coordinating residues labeled. (D) NSP14 in the absence of NSP10 viewed from the same orientation. A hydrophobic cluster adjacent to the active site (shown in Sand color) flips H188 into a position where it likely disrupts the binding of M2 (shown for reference as semitransparent green spheres).
Key insights for anti-SARS-CoV-2 drug development
Structures identified in this paper may serve as exceptional starting point tools for structure-guided development and optimization of specific NSP14 inhibitors, which can be used to tackle COVID-19 and potentially other future viral threats.
“It is important to note that these fragments are soaked into crystals at relatively high concentrations and whilst they may have good ligand efficiency, are not expected to be potent inhibitors without further optimization”, say study authors in this bioRxiv paper.
In any case, this study concludes that druggable MTase domain active site may be the best place for starting the development of broad-spectrum antivirals that may be a valuable addition to our armamentarium against the ongoing COVID-19 pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources