What is furin?
Furin is an endoprotease enzyme that is present in cells and activates several proprotein substrates. A proprotein is a precursor protein that is cleaved by an endoprotease to form the mature and active protein.
Furin is important for embryogenesis, as it catalyzes the maturation of growth factors, receptors, extracellular matrix proteins, and other protease systems. Additionally, furin is a housekeeping protein that is required for several cellular events. Furin is also a prototypic member of the proprotein convertase (PC) family.
Sub-cellular localization and function of furin
Furin resides in the trans-Golgi network (TGN), cell surface, and early endosomes. In the TGN, furin cleaves and activates transforming growth factor β (TGF-β) cytokines, receptors, and viral envelope glycoproteins.
On the cell surface, furin activates proteins involved in cell migration, tumor metastasis, and pathogenic proteins like anthrax protective antigen (PA), proaerolysin, and Clostridium septicum α-toxin. Within the mildly acidic endosomal compartments, profurin is activated by autocatalytic processes and activates A/B-type bacterial toxins, including Pseudomonas exotoxin A and Shiga toxin.
Taken together, furin is trafficked between these different processing compartments. Localization of furin to a specific compartment is mediated by binding to clathrin adaptors or phosphorylation of certain serine residues.
Phosphorylated furin binds to a sorting protein that localizes furin to the TGN or a cycling loop between early endosomes and the cell surface. Dephosphorylation of furin traffics this protein from early endosomes to the TGN.
The binding of furin to actin-binding protein filamin-A (ABP-280) localizes it to the cell surface. Hypoxia in cancer cells induces furin expression and redistribution to the plasma membrane, where it enables cell invasion by activating metalloproteases.
Furin cleavage sites in pathogenic viruses
Viruses can infect certain tissues through a phenomenon termed viral tropism. Furin regulates viral tropism.
The fusion protein precursor HA0 of avian influenza viruses is cleaved by furin. Avian influenza viruses that lack the furin site in HA0 infect the intestinal tract. This HA0 mutation leads to the formation of a furin cleavage site, thereby allowing HA0 to be activated by furin present on all cell surfaces. This enables the virus to infect all cells of the bird, thus causing a systemic infection.
When the H5N1 influenza virus, which is the causative agent of the deadly 1997 Hong Kong flu, was analyzed, two mutations within HA0 were found to generate a tandem furin site. This increased its cleavability by furin and viral tropism. Such mutations have been observed in other pathogenic viruses too.
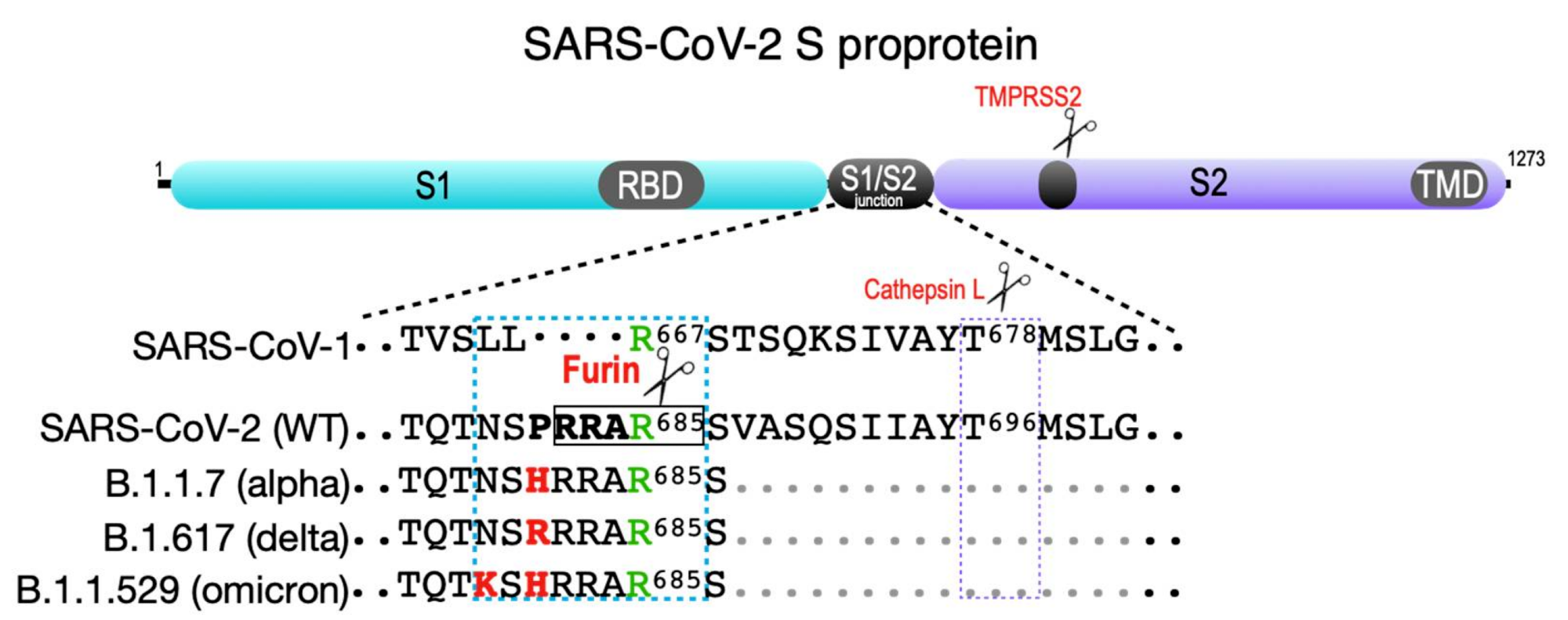 SARS-CoV-2 spike protein. Shown are the S1 and S2 segments in the SARS family S proprotein that flank the S1/S2 cleavage site junction as well as the ACE2 receptor-binding domain (RBD) in S1 and the transmembrane domain (TMD) in S2. The S2′ TMPRSS2 cleavage site is common to all SARS coronaviruses (violet box). The SARS-CoV-1 S1/S2 junction is cut by cathepsin L at Thr678 . SARS-CoV-2 contains a four-amino-acid insertion (PRAA684), which converts the trypsin-sensitive Arg685 residue (green) to the P1 site cut by furin (RRAR685, boxed). The cyan box also shows the B.1.1.7 (alpha variant) furin site containing the P681 → H change; the more transmissible B.1.617.2 (delta variant) furin site, which contains the P681 → R change; and the recently reported B.1.1.529 (omicron variant), which contains both P681 → H and N679 → K changes.
SARS-CoV-2 spike protein. Shown are the S1 and S2 segments in the SARS family S proprotein that flank the S1/S2 cleavage site junction as well as the ACE2 receptor-binding domain (RBD) in S1 and the transmembrane domain (TMD) in S2. The S2′ TMPRSS2 cleavage site is common to all SARS coronaviruses (violet box). The SARS-CoV-1 S1/S2 junction is cut by cathepsin L at Thr678 . SARS-CoV-2 contains a four-amino-acid insertion (PRAA684), which converts the trypsin-sensitive Arg685 residue (green) to the P1 site cut by furin (RRAR685, boxed). The cyan box also shows the B.1.1.7 (alpha variant) furin site containing the P681 → H change; the more transmissible B.1.617.2 (delta variant) furin site, which contains the P681 → R change; and the recently reported B.1.1.529 (omicron variant), which contains both P681 → H and N679 → K changes.
Furin cleavage site in SARS-CoV-2
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has also acquired and optimized the furin cleavage site.
In SARS-CoV and SARS-CoV-2, the spike (S) protein is cleaved by the cell surface protease transmembrane serine protease 2 (TMPRSS2) to expose the receptor-binding domain (RBD). The S protein binds the human receptor, angiotensin-converting enzyme 2 (ACE2) through the RBD; however, TMPRSS2 cleavage does not expose the RBD completely.
Within the S protein is suboptimal furin, whose cleavage by furin fully exposes the RBD and subsequently allows it to efficiently bind ACE2.
The SARS-CoV-2 Alpha variant harbors a P681H mutation in the S protein, which increases cleavability by furin. This change may also trigger cleavage by furin in the endosomal compartments.
Comparatively, the highly transmissible SARS-CoV-2 Delta variant harbors a P681R mutation that increases furin-dependent infectivity. Furthermore, this mutation may also enable cleavage in multiple cellular compartments.
The SARS-CoV-2 Omicron variant contains both P681H and N679K mutations that allow endosomal processing by furin. Whereas the P681H mutation triggers cleavage by furin in the endosomal compartments, the N679K mutation likely further increases cleavage efficiency in endosomes. The N679K mutation also mimics the efficacious compartment-specific furin autoactivation pathway; however, these effects must be proven experimentally.
The maintenance of a suboptimal furin site may allow optimal infectivity of SARS-CoV-2. In fact, the optimization of the furin cleavage site has had debilitating consequences on morbidity and mortality.
Bioinformatic studies have identified polymorphisms in the FURIN gene as risk factors for diabetes, cardiovascular disease, obesity, and all-cause mortality. These risk factors, along with the key role of furin in SARS-CoV-2 pathogenesis, may increase the vulnerability of comorbid patients to severe outcomes of COVID-19.
Furin inhibitors
Since furin is essential for viral pathogenicity, furin inhibitors should be able to block viral infection. This hypothesis has led to the development of an irreversible peptidyl chloromethyl ketone (CMK) active site furin inhibitor.
Cells treated with CMK can block infection by human immunodeficiency virus 1 (HIV-1), paramyxovirus, pathogenic avian influenza viruses, and SARS-CoV-2. However, furin-directed CMKs are non-specific and inhibit other proprotein convertases; thus, these agents are not suitable as therapeutics.
Alternatively, a highly selective furin inhibitor α1-antitrypsin Portland (α1-PDX) has been engineered. To this end, α1-PDX blocks the activation of several pathogenic viruses, including HIV-1, measles virus, and human cytomegalovirus (HCMV).
In cancer models, α1-PDX effectively inhibits tumor cell invasiveness and tumor metastasis. Recombinant α1-PDX reduces atherosclerotic progression, in part by inhibiting the furin-dependent activation of certain metalloproteases.
Currently, there is a lack of expression systems that can generate the physiologically stable α1-PDX. However, several advances have allowed the formulation of α1-PDX and its variants.
For example, donor-purified α1-antitrypsin inhibits the cellular entry of SARS-CoV-2. Thus, this raises the possibility of combining recombinant α1-antitrypsin and α1-PDX for the treatment of COVID-19.
Several cell-penetrant peptide inhibitors have also been developed. To this end, peptidomimetic inhibitors have led to the design of furin inhibitors that prevent pathogen activation at very low concentrations. For example, one study had shown that the small-molecule furin inhibitor BOS-981 blocks the cleavage of the S protein S1/S2 site.
Conclusions
Selective furin inhibitors may provide a broad-based therapeutic option for the treatment of multiple diseases, including COVID-19.
Journal reference:
- Thomas, G., Couture, F., & Kwiatkowska, A. (2022). The Path to Therapeutic Furin Inhibitors: From Yeast Pheromones to SARS-CoV-2. International Journal of Molecular Sciences 23(7). doi:10.3390/ijms23073435.