The SARS-CoV-2 S protein
The SARS-CoV-2 S protein is the most studied structural protein of SARS-CoV-2, as it helps in target recognition and cellular entry that facilitates viral infection. As a result, the S protein has been the primary target for the evolution of SARS-CoV-2 variants of concern (VOC) such as the Alpha, Beta, Gamma, Delta, and Omicron variants.
The evolution and higher infectivity of SARS-CoV-2 VOCs are attributed to mutations that have occurred rapidly in the S protein over time. The D614G mutation, which was first identified in the middle of 2020, has been identified in all major SARS-CoV-2 VOCs and plays a major role in the increased infectivity of these strains.
The SARS-CoV-2 Omicron variant has a total of 34 mutations in its S protein as compared to the wild-type SARS-CoV-2 S protein. Given the rapid rate at which several mutations in the S protein have emerged, it has become increasingly important to understand the mechanisms behind the development of these mutations and their clinical impact on new infections.
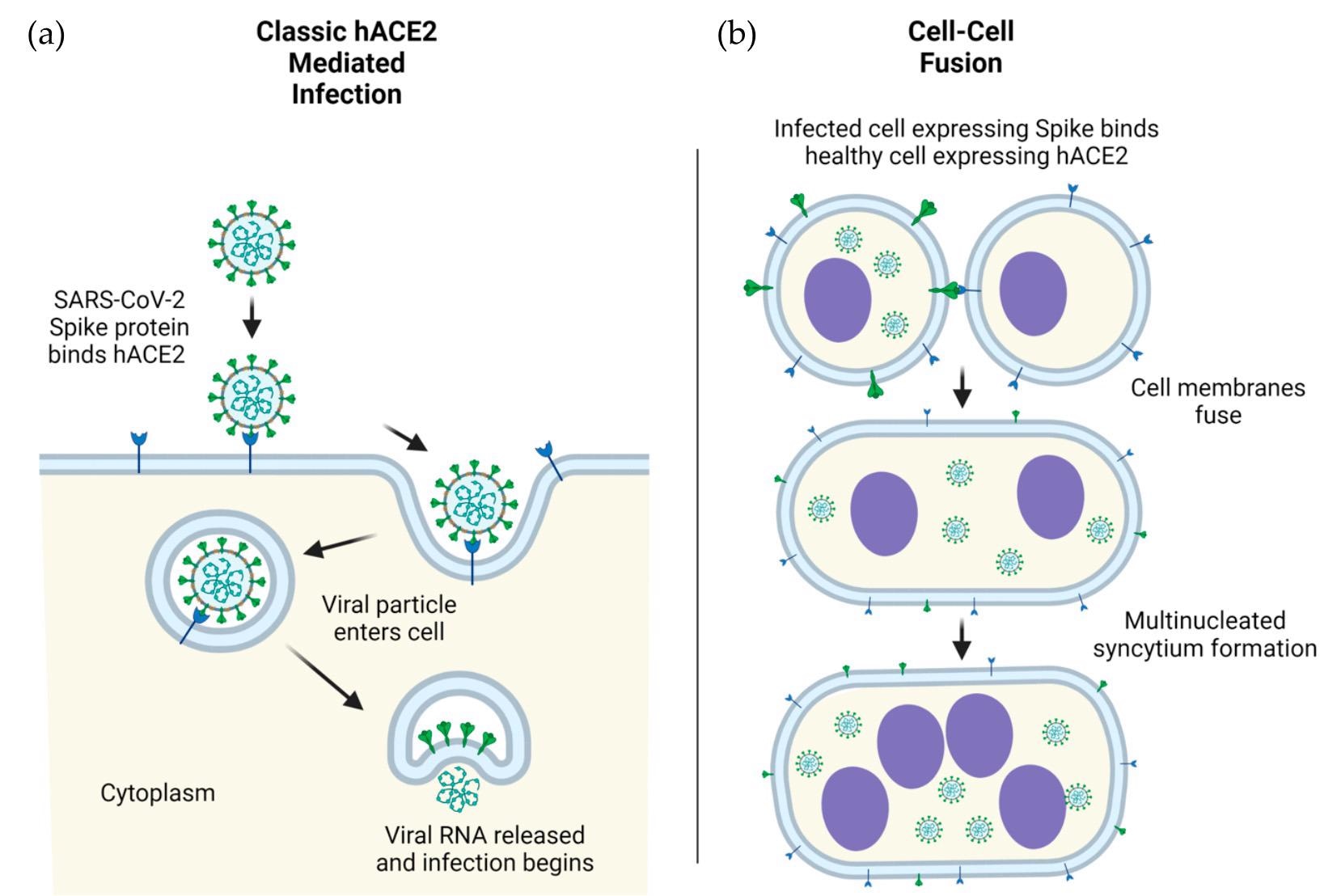
SARS-CoV-2 viral particles into human cells, mediated by ACE2. (b) Cellular infection by ACE2- spike mediated cell-cell fusion. Infection in human (h) cells is used as an example. Figure 1. ACE2-mediated cellular infection by SARS-CoV-2. (a) Schematic of direct cellular entry of SARS-CoV-2 viral particles into human cells, mediated by ACE2. (b) Cellular infection by ACE2-spike mediated cell-cell fusion. Infection in human (h) cells is used as an example.
S protein structure and function
The SARS-CoV-2 S protein consists of two S1 and S2 subunits. The S1 subunit, which is further subdivided into an N-terminal domain (NTD) and C-terminal receptor-binding domain (RBD), is responsible for target recognition and binding. Comparatively, the S2 subunit consists of a fusion peptide (FP), two heptad repeats (HR1 and HR2), a transmembrane, and a C-terminal tail and plays a role in membrane fusion and endosomal escape.
The mechanism of SARS-CoV-2 infectivity is initiated by the RBD, as it allows for viral entry by recognizing the angiotensin-converting enzyme 2 (ACE2) receptor. Comparatively, the NTD assists in S protein structural conformation and, as a result, plays a significant role in the immune escape capabilities of the virus. After viral binding attachment through host ACE2, the FP, HR1, and HR2 subdomains form a conformation that allows entry of SARS-CoV-2 into the host.
The C-terminal subdomain of the S2 promotes the escape of the S protein from the endoplasmic reticulum (ER), thereby causing aggregation of S proteins on the surface. This S protein aggregation helps in binding to other ACE2 receptors present on the healthy cells, subsequently leading to widespread infection.
![Mutations within the NTD of SARS-CoV-2 spike protein. (a) Structural representation of substitutions (red) within the NTD (green) of SARS-CoV-2 spike protein. (b) Visual representation of mutations within the NTD of spike protein. (c) Frequency, residue distance from NTD “supersite”, modification in charge at physiological pH, and change in hydrophobicity at pH 7. Residue distance was calculated in PyMOL on the protein three-dimension structure (PDB-6ZGG), by measuring the distance between the nearest atom of “supersite” amino acids identified by Mccalum et al., 2021 [32] and the nearest atom of amino acids of interest.](https://www.news-medical.net/images/news/ImageForNews_709293_16487727573881890.jpg) Mutations within the NTD of SARS-CoV-2 spike protein. (a) Structural representation of substitutions (red) within the NTD (green) of SARS-CoV-2 spike protein. (b) Visual representation of mutations within the NTD of spike protein. (c) Frequency, residue distance from NTD “supersite”, modification in charge at physiological pH, and change in hydrophobicity at pH 7. Residue distance was calculated in PyMOL on the protein three-dimension structure (PDB-6ZGG), by measuring the distance between the nearest atom of “supersite” amino acids identified by Mccalum et al., 2021 [32] and the nearest atom of amino acids of interest.
Mutations within the NTD of SARS-CoV-2 spike protein. (a) Structural representation of substitutions (red) within the NTD (green) of SARS-CoV-2 spike protein. (b) Visual representation of mutations within the NTD of spike protein. (c) Frequency, residue distance from NTD “supersite”, modification in charge at physiological pH, and change in hydrophobicity at pH 7. Residue distance was calculated in PyMOL on the protein three-dimension structure (PDB-6ZGG), by measuring the distance between the nearest atom of “supersite” amino acids identified by Mccalum et al., 2021 [32] and the nearest atom of amino acids of interest.
S protein mutations
The characteristics associated with SARS-CoV-2 VOCs, according to the World Health Organization (WHO), include their increased transmissibility and varied disease presentation. Mutations in the S protein play a critical role in the increased fitness of these variants.
Mutations in the RBD can significantly affect the binding of the S protein to the ACE2 receptor, thus altering the infectivity of variants harboring these mutations. Recently, researchers found that instead of the binding affinity of the S protein to ACE2 being the primary factor for the positive selection of mutants, neutralization resistance plays a more dominant role. Similarly, another study suggested that mutations in the RBD cause changes in its hydrophobicity and charge, thus leading to increased immune escape.
Within the Omicron variant, 13 RBD mutations have been identified, of which ten have been detected in the defining sequence. In one recent study, researchers postulated that the five defining mutations of K417, E484, Q493, Q498, and N501 within the Omicron RBD may have arisen in murine species before returning to humans.
For the NTD domain, neutralizing antibodies target the antigenic “supersite,” thus contributing to positive selection for SARS-CoV-2. Two NTD mutations of T95I and ∆69–70 are considered to be responsible for the increased fitness and infectivity of the Omicron variant, which is likely due to the remodeling of the supersite by these mutations.
Aside from the S1 subunit, mutations that occur between the S1 and S2 subunits also influence the infectivity and fitness of a SARS-CoV-2 variant. The D614G mutation, for example, occurs outside the defined domains that alter RBD conformation and has been shown to enhance the binding affinity between the S protein and ACE2. Additionally, the P681 mutation, which occurs in the 680-689 amino acid stretch, increases the furin cleavage responsible for greater infectivity and disease severity in certain variants.
Other mutations of interest
Four S2 subunit mutations including T716I, D950N, S982A, and D1118H occur commonly in SARS-CoV-2 variants, except for the Omicron variant. Additionally, the D950N and S982A mutations in the heptad repeats HR1 may indicate their role in the positive selection of variants. Furthermore, the S982A mutation appears to increase the presentation of the “up” RBD state by preventing the interaction with T547, thereby stabilizing the “down” RBD state.
Conclusions
The current review article provides insight into the potential mechanisms that lead to common mutations of the S protein found in SARS-CoV-2 VOCs that enhance their fitness. The findings discussed here highlight the importance of studying SARS-CoV-2 mutations, their evolution, and potential therapies that could target particularly harmful mutations.
Journal reference:
- Magazine, N., Zhang, T., Wu, Y., et al. (2022) Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses. doi:10.3390/v14030640.