The current coronavirus disease 2019 (COVID-19) pandemic, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains a major threat to global health. The most common symptom associated with COVID-19 is respiratory distress; however, this disease can cause a wide range of symptoms.
Background
Recent evidence has shown that SARS-CoV-2 has the potential to infect the human central nervous system (CNS), resulting in neuroinflammation. The association between COVID-19 and neurological complications has raised concerns about its potential to impact the development of certain neurodegenerative diseases like Parkinson’s disease (PD), Alzheimer’s disease (AD), and amyotrophic lateral sclerosis (ALS), all of which have been linked to certain viral infections.
To further investigate the relationship between SARS-CoV-2 infection and neurodegenerative diseases, researchers in a recent Protein & Cell study discover that SARS-CoV-2 alters the disassembly of stress granule (SG) proteins, thus stimulating their amyloid aggregation in a pattern that resembles ALS-associated aggregation.
Does SARS-CoV-2 need ACE2 for entry?
Both Vero cells, which naturally express the angiotensin-converting enzyme 2 (ACE2) receptor that is primarily used by SARS-CoV-2 for viral entry, and HeLa cells lacking ACE2 were infected with SARS-CoV-2 for 30 minutes.
Despite lacking the ACE2 receptor, HeLa cells exposed to a high multiplicity of infection (MOI) of 0.75 were still able to be infected by SARS-CoV-2 and retain expression of the viral nucleocapsid (N) protein. HeLa infected cells did not retain expression of the viral (S) spike protein, which suggests poor viral replication in this cell line. However, when HeLa cells were engineered to overexpress ACE2, SARS-CoV-2 infection successfully led to the expression of viral proteins for two days after the initial infection.
Taken together, these initial experiments demonstrate that although ACE2 is not required for the entry of SARS-CoV-2 into cells, it may increase the efficiency of viral infection.
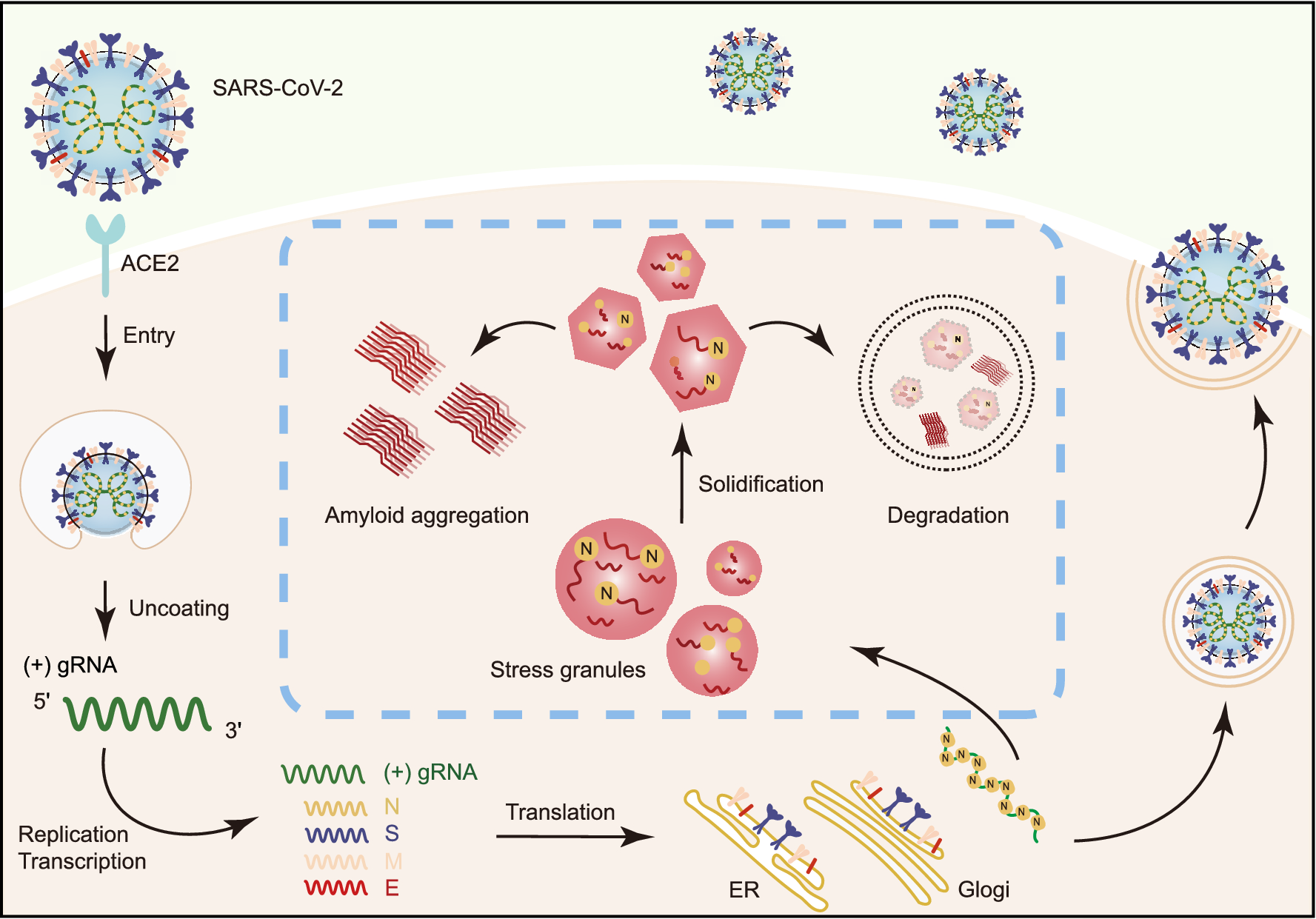 Schematic diagram for the interplay between SARS-CoV-2 and host SGs. During SARS-CoV-2 replication in host cells, the viral N protein enters the host SGs and directly interacts with SG-related amyloid-forming proteins (e.g., FUS, hnRNPA1 and TDP43), which stimulates the liquid to solid phase transition (amyloid formation) of these host proteins. Inefficient maintenance of proteostasis may result in accumulation of the pathological amyloid fibrils and development of neurodegeneration
Schematic diagram for the interplay between SARS-CoV-2 and host SGs. During SARS-CoV-2 replication in host cells, the viral N protein enters the host SGs and directly interacts with SG-related amyloid-forming proteins (e.g., FUS, hnRNPA1 and TDP43), which stimulates the liquid to solid phase transition (amyloid formation) of these host proteins. Inefficient maintenance of proteostasis may result in accumulation of the pathological amyloid fibrils and development of neurodegeneration
Entry of SARS-CoV-2 N protein into SGs
To produce SGs, infected cells were challenged with 100 mol/L sodium arsenite. This was followed by immunofluorescence microscopy to monitor the localization of SARS-CoV-2 N and S proteins, as well as non-structural protein 1 (Nsp1) and Nsp8, as well as open-reading frame 7 (ORF7).
The only SARS-CoV-2 protein capable of entry into the SGs was the N protein, which confirmed previous studies demonstrating that SARS-CoV-2 can invade SGs due to overexpression of N protein.
The impact of the SARS-CoV-2 N protein on SG dynamics
Before relieving infected cells of stress by washing out the sodium arsenite, similar amounts of SGs were identified in both SARS-CoV-2 infected and control cells. After one hour of being washed out, the researchers found that both HeLa and ACE2-overexpressed HeLa cells previously infected with SARS-CoV-2 exhibited a reduced rate of SG disassembly. Notably, HeLa cells that overexpressed the SARS-CoV-2 N protein alone also exhibited a similar level of reduced SG disassembly, thus confirming the role of the N protein in altering SG dynamics.
When both SARS-CoV-2-infected and control cells experienced additional stress for five hours, a greater level of SG disappearance was observed in infected cells. This level of severe SG disappearance was also similar in HeLa cells that overexpressed the N protein, thus providing additional evidence for the role of the SARS-CoV-2 N protein in abnormal SG behaviors.
Previous studies have demonstrated that the liquid-liquid phase separation (LLPS) ability of the SARS-CoV-2 N protein can impact viral genome packing in other viruses. In the current study, the researchers confirmed this finding when the N protein rapidly underwent LLPS in the presence of synthetic single-stranded ribonucleic acid (polyU).
Like the SARS-CoV-2 N protein, several RNA-binding proteins of SGs including fused in sarcoma (FUS), heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), and TAR DNA binding protein 43 (TDP43) are also associated with a high potential of LLPS. Thus, the researchers of the current study were interested in determining the impact of the SARS-CoV-2 N protein on these proteins.
Consistent with previous reports, the researchers confirmed that the N protein can co-phase separate with FUS, TDP43, and hnRNPA2 in LLPS droplets. Furthermore, incubation of the N protein with FUS protein for 12 hours altered the shape of the droplets to be less round than their baseline appearance, while also exhibiting reduced mobility and an inability to recover after bleaching. Similar alterations in the liquid-like nature of the LLPS droplets were observed when the N protein was incubated with hnRNPA1 and TDP43.
The low-complex (LC) domains of the FUS, hnRNPA1, and TDP43 are considered to be the amyloid-forming core sequences of these proteins. Incubation of the SARS-CoV-2 N protein with these domains was found to significantly enhance thioflavin T (ThT) fluorescence intensities, which correlates with amyloid fibril formation. This effect of the SARS-CoV-2 N protein was also confirmed through transmission electron microscopy (TEM).
Effects of SARS-CoV-2 infection on ALS-mutant cells
The researchers then utilized a cell model that overexpresses FUS with the P525L mutation, which impairs the nuclear localization of FUS and subsequently leads to its overaccumulation in the cytoplasm. Following SARS-CoV-2 infection of these cells, the viral N protein was found to co-localize with FUS P525L aggregates.
As compared to control cells, FUS P525L aggregation significantly increased considerably after SARS-CoV-2 infection. Similar aggregation of FUS P525L was found with SARS-CoV-2-infected HeLa cells, with or without the overexpression of SARS-CoV-2 N protein.
Implications
The current study found that HeLa cells, which lack the ACE2 receptor-like neurons, are vulnerable to SARS-CoV-2 infection. These findings align with recent studies that have discovered various other cellular receptors involved in SARS-CoV-2 cell entry, thus confirming the ability of this virus to enter the brain.
Disturbance of SG dynamics has been linked to neurodegenerative conditions like ALS. The current study confirms that SARS-CoV-2 infection, particularly through activities mediated by the N protein, impairs SG dynamics and promotes amyloid aggregation of SG-related proteins.
Taken together, these data demonstrate that SARS-CoV-2 infection has the potential to contribute to neurodegeneration. Further research is therefore needed to understand whether neurodegeneration is, in fact, a long-term consequence of COVID-19.