The human ACE2 protein contains eight cysteine residues, six of which are present in three pairs of disulfide bonds. The remaining two (Cys261 and Cys498) are free thiols and thus available for S-nitrosylation of ACE2 via a reversible nucleophilic attack on nitroso nitrogen to form SNO-protein adducts.
Aminoadamantane nitrate compounds such as amantadine and memantine are non-toxic and have demonstrated robust in vitro and in vivo activity. However, evidence of their efficacy against SARS-CoV-2 in humans is lacking.
Study: Targeted protein S-nitrosylation of ACE2 as potential treatment to prevent spread of SARS-CoV-2 infection. Image Credit: Design_Cells / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the present study, researchers developed aminoadamantane nitrates, which they believed would prove a novel mechanism for therapeutic action against coronavirus disease 2019 (COVID-19). They investigated the molecular mechanism of NO in inhibiting SARS-CoV-2. The authors built upon their previous experience in developing the aminoadamantane drug memantine for treating Alzheimer’s disease.
Human embryonic kidney cells (HEK293T) and Henrietta Lacks (HeLa) cell cells were cultured and mutant ACE2 constructs (with C498A/C262A mutations) were generated. Aminoadamantane nitrate compounds (coded as NMT1 (memantine), NMT2, NMT3, NMT4 (amantadine), NMT5 to NMT9, and NMT5-Met (metabolite, sans nitro group) were produced. The biotin-switch assay was performed for analyzing the S-nitrosylated proteins. In addition, the SARS-CoV-2 S-exposed cells were subjected to immunochemistry analysis. The N-terminal peptidase domain of human ACE2 was cloned and fused with C-terminal (His-tag), and the His-tagged ACE2 protein was then purified.
The cultured cells were co-immunoprecipitated and subjected to immunoblotting with anti-ACE2 and anti-SARS-CoV-2 S antibodies. Mass spectrometry analysis of the S-nitrosylated ACE2 protein was performed and subsequently, the glycosylated and S-nitrosylated ACE2/receptor-binding domain (RBD) construct was prepared and subjected to molecular dynamics (MD) simulations. In addition, patch-clamp analysis and SNO-ACE2 targeting assays were performed to assess the inhibition of viroporin channels.
For pharmacokinetic analyses, eight-week-old Golden Syrian hamsters were challenged intranasally with 1 × 105 or 106 plaque-forming units (PFU) of SARS-CoV-2 (USA-WA1/2020 strain). Subsequently, they were administered two doses of the nitrate compounds by oral gavage 12 hours apart, following which their blood samples and lung samples were collected for determining viral titers and histological changes using plaque assays and immunohistochemistry, respectively. Remdesivir, apilimod, and puromycin were used as positive controls.
The selectivity index (SI) was calculated to determine the therapeutic potential of the compounds and compares the compound’s half-maximal non-specific cytotoxicity (CC50) to its half-maximal effective antiviral concentration (EC50) (CC50/EC50). The team also determined the maximally tolerated dose (MTD) of NMT3 and NMT5 in vivo based on dose-ranging toxicity and efficacy studies in Syrian hamsters.
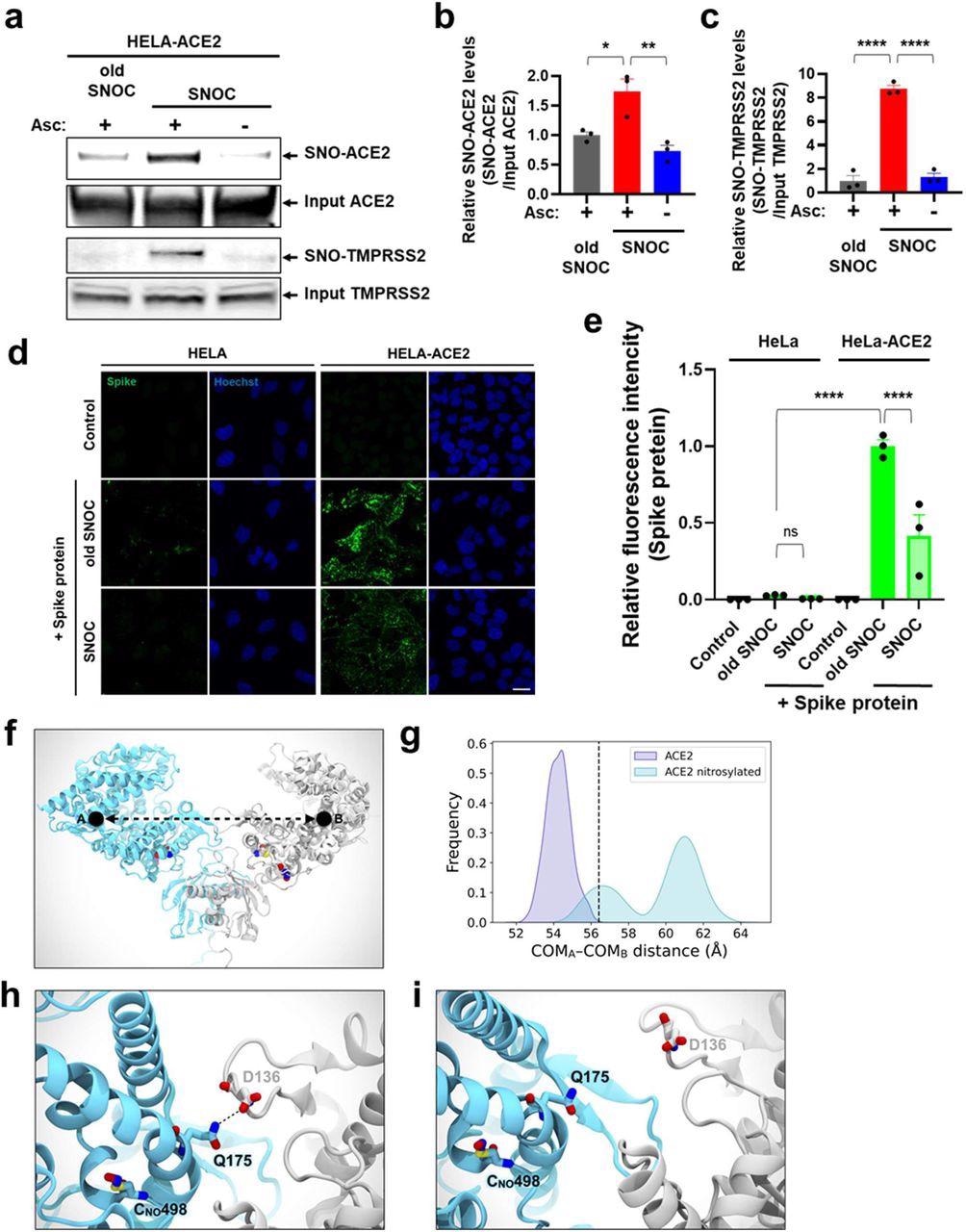
SNOC increases S-nitrosylation of ACE2 and inhibits binding of SARS-CoV-2 Spike (S) protein.a, Assay for SNO-ACE2 and SNO-TMPRSS2 in HeLa-ACE2 cells. Cells were exposed to 100 μM SNOC or, as a control, ‘old’ SNOC (from which NO had been dissipated). After 20 minutes, cell lysates were subjected to biotin-switch assay to assess S-nitrosylated (SNO-) and input (total) proteins detected by immunoblotting with cognate antibody. The ascorbate minus (Asc-) sample served as a negative control. b, c, Ratio of SNO-ACE2/input ACE2 protein and SNO-TMPRSS2/input TMPRSS2 protein. Data are mean + s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA with Tukey’s multiple comparisons. n = 3 biological replicates. d, HeLa and HeLa-ACE2 cells were pre-exposed to 100 μM SNOC or old SNOC. After 30 minutes, 10 μg/ml of purified recombinant SARS-CoV-2 Spike (S1+S2) protein was incubated with the cells. After 1 h, cells were fixed with 4% PFA for 15 minutes, and bound Spike protein was detected by anti-Spike protein antibody; nuclei stained with 1 μg/ml Hoechst. Cells were imaged by confocal fluorescence microscopy. Scale bar, 20 μm. e, Quantification of relative fluorescence intensity. Data are mean + s.e.m., ****P < 0.0001 by ANOVA with Tukey’s multiple comparisons. n = 3 biological replicates. f, Molecular representation of the S-nitrosylated-ACE2/RBD model upon transient detachment at the level of the peptidase domain dimeric interface. SNO-Cys261 and SNO-Cys498 are shown with Van der Waals spheres. The black dots indicate qualitative placement of centers of mass (COM) for each ACE2 protomer, and the dashed arrow represents the distance between COMs. Spike’s RBDs and N-glycans, which were included in the simulation, are hidden for image clarity. SpBD, Spike binding domain; CLD, collectrin-like domain; PD, peptidase domain. g, Distribution of the distance between COMs from molecular dynamics simulations of WT ACE2/RBD (purple) vs. nitrosylated-ACE2/RBD (cyan). Dashed black line at approximately 56.5 Å indicates the reference distance between COMs calculated from the cryo-EM structure (PDB: 6M17). S-Nitrosylated-ACE2/RBD shows an overall larger distance between COMs with a bimodal distribution. h, Close-up image illustrating Q175A to D136B interaction present in starting conformations of the S-nitrosylated-ACE2 system. i, Close-up image illustrating the disruption of the interaction between Q175A and D136B occurring along the dynamics of the S-nitrosylated-ACE2 system.
Results
Among the seven aminoadamantane nitrates tested, NMT5 demonstrated the highest stability and therapeutic potential (SI = 9.2, EC50 5.3 μM) at concentrations attainable in human tissues at well-tolerated doses. In addition, NMT3 (NitroSynapsin), also showed some protection against SARS-CoV-2 (EC50 87.7 μM). Thus, these two compounds were used for further analyses.
NMT3- or NMT5- mediated SNO-ACE2 formation could suppress SARS-CoV-2 infection by ancestral strain and Delta and Omicron variants by about 95%. NMT5 could modify ACE2 at both of the cysteine residues (Cys261 and Cys498), which were susceptible to S-nitrosylation by S-nitrosocysteine (SNOC). ACE2 was successfully S-nitrosylated by SNOC. A significantly decreased binding of purified S protein with SNOC was observed, indicating that cysteine residues were susceptible to S-nitrosylation.
In the bio-switch assays, the cysteine residues Cys261 and Cys498 of C261A/C498A mutation significantly inhibited SNOC-mediated S-nitrosylation, indicating these two residues are S-nitrosylation targets. Mass spectrometry analyses confirmed this finding. NMT5 preferentially S-nitrosylated Cys498 over Cys261.
In the co-IP experiments, NMT5 (5 µM) significantly decreased the protein immunoprecipitation, indicating that the compounds inhibited S protein-ACE2 binding. In the plaque assays, NMT5 (but not NMT3), substantially reduced viral titers by ~100 fold. In the histological examination, NMT5 eliminated COVID-19-related pulmonary hemorrhages when examined five days post-infection. In the immunohistochemistry analysis, NMT 5 reduced the proinflammatory cytokine and chemokine expression.
NMT5 inhibited viral entry in a dose-dependent fashion, with 5 µM inhibiting 53%, 10 µM inhibiting 76%, and 20 µM inhibiting 92%. NMT3 showed a more limited ability to suppress pseudovirus entry, 24% at 10 µM. NMT5 failed to S-nitrosylate other proteins, such as TMPRSS2, S, or E protein, demonstrating the selectivity of NMT5 for ACE2. The half-life in plasma for NMT3 and NMT5 were 7.9 hours and 10.6 hours, respectively. The mean Cmax for NMT5 and NMT3 were 0.2 µM and 0.4 µM, respectively.
The authors found that mechanistically, NMT5 would bind to the E protein viroporin channel on SARS-CoV-2 and subsequently transfer the NO group to ACE2 on the host cell to prevent infection.
Overall, the study findings demonstrated that the cellular receptor of SARS-CoV-2 entry, ACE2, can be S-nitrosylated to inhibit the S-ACE2 binding and thereby inhibiting viral replication and infectivity. Additionally, NMT5, a novel aminoadamantane nitrate developed by the authors demonstrated potent SARS-CoV-2 inhibition by S-nitrosylation with a nitro group that blocked the E protein formed- viroporin channels.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Targeted protein S-nitrosylation of ACE2 as a potential treatment to prevent the spread of SARS-CoV-2 infection, Chang-ki Oh1, Tomohiro Nakamura1, Nathan Beutler2, Xu Zhang1, Juan Piña-Crespo1, Maria Talantova1, Swagata Ghatak1, Dorit Trudler1, Lauren N. Carnevale1, Scott R. McKercher1, Malina A. Bakowski3, Jolene K. Diedrich1, Amanda J. Roberts4, Ashley K. Woods3, Victor Chi3, Anil K. Gupta3, Mia A. Rosenfeld5, Fiona L. Kearns5, Lorenzo Casalino5, Namir Shaabani2, Hejun Liu6, Ian A. Wilson6, Rommie E. Amaro5, Dennis R. Burton2, John R. Yates III1, Cyrus Becker7, Thomas F. Rogers2,8, Arnab K. Chatterjee3, Stuart A. Lipton, bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.04.05.487060, https://www.biorxiv.org/content/10.1101/2022.04.05.487060v1
- Peer reviewed and published scientific report.
Oh, Chang-ki, Tomohiro Nakamura, Nathan Beutler, Xu Zhang, Juan Piña-Crespo, Maria Talantova, Swagata Ghatak, et al. 2022. “Targeted Protein S-Nitrosylation of ACE2 Inhibits SARS-CoV-2 Infection.” Nature Chemical Biology, September. https://doi.org/10.1038/s41589-022-01149-6. https://www.nature.com/articles/s41589-022-01149-6.