The ongoing coronavirus disease 2019 (COVID-19) pandemic has not only resulted in global public health crises but has also created havoc in the day-to-day lives of people across. Treatments targeting the causative novel coronavirus (severe acute respiratory disease coronavirus 2 – SARS-CoV-2) infection are mainly supportive and provide symptomatic relief.
Development of newer therapies targeting the pathogenesis of the viral infection through gaining a deeper understanding of the SARS-CoV-2-to-host interaction is warranted.
Alternative splicing (AS) – a mechanism that regulates proteome diversity through single RNA-splicing, generates alternative mRNAs that encode distinct protein isoforms. This pathway can be hijacked by numerous viruses for enabling their replication and host-immune evasion. In the SARS-CoV-2, Ddx58 protein attaches to the cellular spliceosome complex to inhibit splicing.
Contrastingly, it induces numerous AS in the host RNAs, rendering alternative proteins, thus hampering the cell cycle, DNA synthesis, and immune responses.
However, whether AS is instrumental in the pathogenesis of COVID-19 remains obscure. This can be uncovered by characterizing the splicing landscape in host cells after SARS-CoV-2 infection.
 Study: Abnormal global alternative RNA splicing in COVID-19 patients. Image Credit: NIAID
Study: Abnormal global alternative RNA splicing in COVID-19 patients. Image Credit: NIAID
The study
This study entailed the integration of multiple-omics datasets to examine the dysregulation in the host splicing mechanism and alteration in protein isoforms among COVID-19 patients.
The goal of this study published in the journal PLOS Genetics was to explore the impact of an altered AS landscape on clinical outcomes and identify potential transformations in the drug-binding sites for proteins in COVID-19 patients.
Histological samples from the lungs of nine COVID-19 fatalities in the early outbreak from two hospitals in Wuhan, China, were selected. Healthy lung tissue samples of lung cancer patients served as matched controls
Findings
Of the nine selected COVID-19 lung samples, five were from male patients; the mean age of the patients was 68 years. The average symptomatic period of these patients was ~26 days before death.
A standard approach for treating COVID-19 did not exist during the initial outbreak—the phase of occurrence of these fatalities. Therefore, these patients were given different antimicrobials and immunomodulatory agents.
It was observed that all infected samples had inflammatory infiltrates in varying degrees – predominantly myeloid cells and lower proportions of T and B cells. Eight infected samples exhibited lower amounts of epithelial cells. A single-cell RNA sequencing (scRNA-seq) group showed an identical distribution of inflammatory cell types.
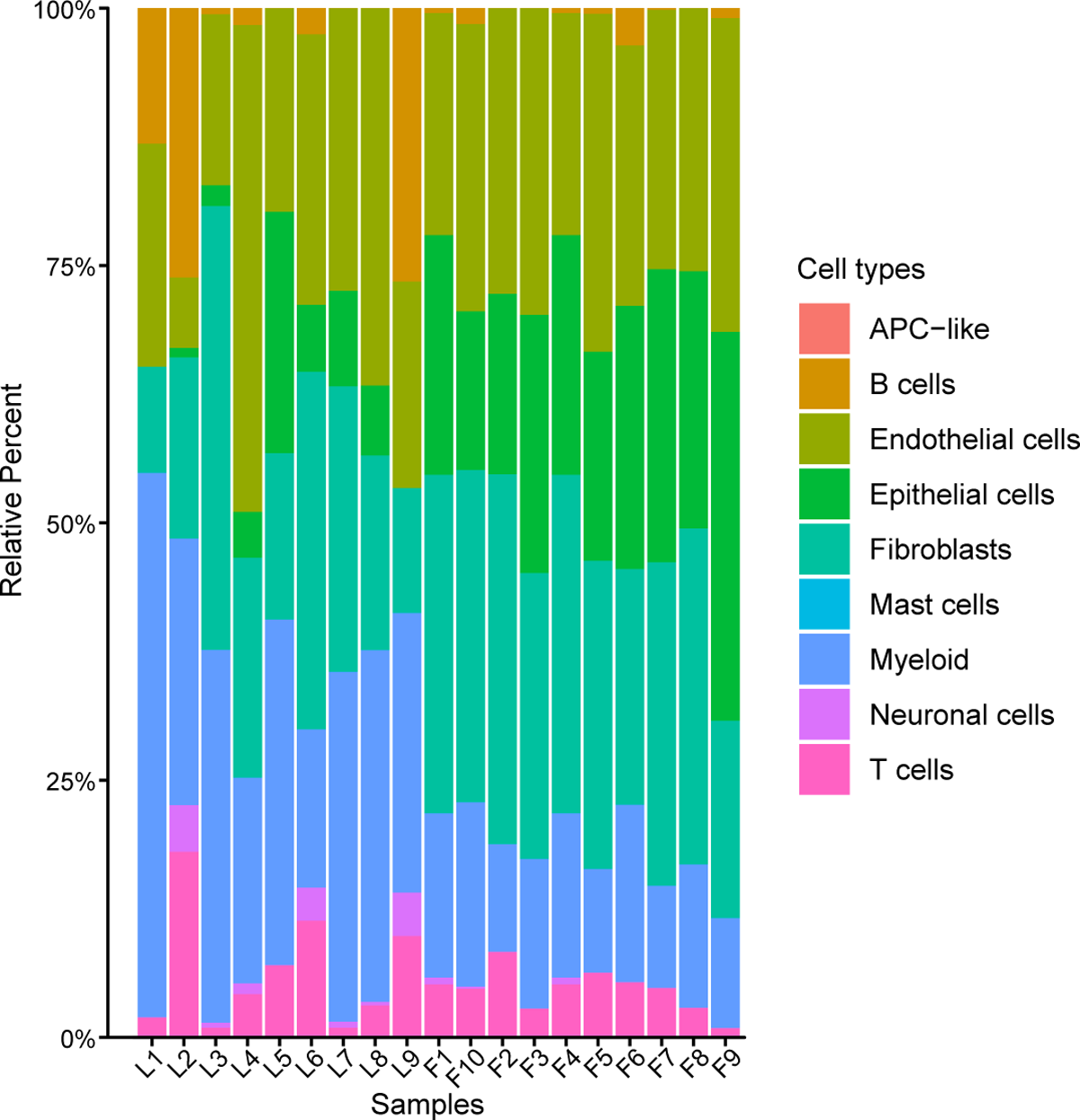
The proportion of 9 cell subpopulations in 9 COVID-19 and 10 control samples. The colors indicate cell type information. L1-L9, COVID-19 patients. F1-F10, control samples.
Proteomic analysis on the SARS-CoV-2 infected lung samples revealed 4,689 proteins. Among these, 235 upregulated whereas 402 downregulated proteins in COVID-19 patients. In addition, gene ontology (GO) enrichment analysis detected dysregulation of the neutrophil process in infected patients with severe disease. These processes included – coagulation and hemostasis.
Investigation of the virus-host protein-protein interaction (PPI) network suggested profound dysregulation in the host splicing machinery in the infected samples – which is likely to alter the AS of the genes and thus, may impact the host immune response.
Bulk RNA sequencing identified 1,383 genes; the major transcript showed changes in the differential transcript usage (DTU) in COVID-19 patients. In fact, multiple DTU genes were instrumental in RNA splicing – ten genes escalated major transcript usage while 18 others decreased it in the infected patients.
Meanwhile, the lung parenchyma displayed only a few cytokine genes with varying major transcript usage. Of note, the crucial role of interleukin (IL)2 in T-cell proliferation and effector and memory T-cell generation was recognized. The results indicated that the IL2 transcript switch might affect T-cell activation in patients with severe disease.
Overall, 3,937 differential transcript expression (DTE) were observed. In patients who died, 1,890 DTEs were upregulated while 2,047 were down-regulated. A majority of the genes with upregulated transcripts were associated with oxidative stress response – signified by intense breathing difficulties in severe COVID-19 disease.
The findings indicated that SARS-CoV-2 recontours the central cellular pathways such as translation and carbon metabolism. Additionally, patients who died were speculated to harbor a dysregulation of bidirectional interactions of the lung and brain.
Exon skipping (ES) denoted the most common local splicing change, followed by complex event; intron retention; alternative 3’ splice site; and alternative 5’ splice site. The functional consequences on protein isoforms were predictable at numerous AS events.
A great divergent co-splicing connections network was detected amongst the case and control groups. It was noted that the DDX3X – the neutrophil activation network hub, encodes a DEAD-box RNA helicase while the 18th module exhibits a greater capacity for macrophage activation in COVID-19 patients.
There existed a continuous dysregulation of the differential expression transcripts amongst moderate to ICU stages of the infected patients. In addition, the disease groups and healthy donors expressed pervasive DTU and DTE. The results suggested that SARS-CoV-2 triggers comprehensive dysregulation in transcriptions of the host immune cells.
It was also observed that the elevated DTE numbers correlated to worse clinical outcomes – hence, COVID-19 patients can be deemed a different transcriptomic phenotype compared to non-infected individuals. Furthermore, alternative transcript usage rendered marked phase specificity of the disease.
The findings hinted toward the pivotal role of coagulation signature disruptions in COVID-19 exacerbations. It was also emphasized that the host immunity puts up an efficient antiviral response during the early phases of the infection, especially In patients with severe symptoms – evident through the upregulated neutrophil signature in severe and ICU cases.
Widespread AS events were detected in samples of COVID-19 patients irrespective of the disease severity—most were ES events. AS of LRRFIP1 was found to be a prevalent signature in patients with severe disease. Hence, a strong correlation was found between the COVID-19 severity and the transcript isoform changes in PBMC samples of the infected patients.
A markedly higher usage of isoform 017—which is crucial for PPARG stimulation by drugs—was detected. The isoform 002 showed a more robust interaction compared to isoform 017 with telmisartan molecules. These alterations led to the inadequate immunomodulatory therapy response in patients with severe disease. It was stated that the canonical isoform 002 shows increased usage in normal tissues and could aid in mitigating the cytokine storm induced by SARS-CoV-2 infection.
The results underscored that during widespread AS regulation after SARS-CoV-2 infection, targets for altered splicing are specially chosen for facilitating pathogen survival. Moreover, exon-specific AS detection could be a reliable biomarker and serve as a novel approach in the diagnosis and monitoring of COVID-19 progression.
The findings provided additional insights into the complexity of RNA splicing upon SARS-CoV-2 infection—which could enable COVID-19 diagnosis and therapy.