In a recent study posted to the bioRxiv* preprint server, researchers reported that nanoparticles could prevent severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) cell entry.
 Study: ACE2 nanoparticles prevent cell entry of SARS-CoV-2. Image Credit: Kateryna Kon / Shutterstock
Study: ACE2 nanoparticles prevent cell entry of SARS-CoV-2. Image Credit: Kateryna Kon / Shutterstock

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
SARS-CoV-2, the causal pathogen of coronavirus disease 2019 (COVID-19), enters human cells by interacting through its spike (S) protein with the host angiotensin-converting enzyme-2 (ACE2). A number of vaccines and treatment approaches have been developed against SARS-CoV-2. These options are mostly based on antibodies that bind to exposed viral proteins and prevent their entry into cells.
Among the concerns with these approaches is the evolution of the virus into novel mutant variants that weaken the interaction between the virus and the antibody, thereby reducing the effectiveness of vaccines and therapeutics. For instance, the latest Omicron variant, with around 15 mutations in the S protein’s receptor-binding domain (RBD), exhibits substantial immune escape and negatively affects the vaccination or therapeutic efficiency.
Furthermore, some therapeutics target proteins that are involved in viral replication. Moreover, since these inhibitory mechanisms bind to proteins that are potentially subject to mutations, they could also allow the virus to escape. In addition, drugs that target the replication process might also cause complications since their functional activity takes place inside the host cell rather than blocking viral entry. Thus, it is critical to explore the ACE2 receptor as a therapeutic intervention and a barrier to viral escape.
The study and findings
In the present study, researchers engineered a decoy or nanoparticle, a murine leukemia virus (MLV)-based carrier system pseudo-typed with full-length human ACE2 (hACE2) on the surface. In parallel, MLVs with luciferase messenger ribonucleic acid (mRNA) were established as control MLVs together with other controls like MLVs pseudo-typed with S proteins of wildtype (WT) SARS-CoV-2, Delta, or Omicron variant. The effect of hACE2 nanoparticles on cell entry of SARS-CoV-2 was studied using luciferase activity assays.
The morphology and presence of pseudoviruses post-infection of Vero-E6 cells were verified using cryogenic electron tomography (cryo-ET). These cells endogenously express ACE2 of African green monkeys with a 100% sequence identity with hACE2. The authors observed S proteins projecting from the pseudo viral surface in pre- and post-fusion configurations. They reported that pseudo viral cell entry was based on clathrin-mediated endocytosis, consistent with SARS-CoV-2 cell entry in the absence of transmembrane serine, protease 2 (TMPRSS2).
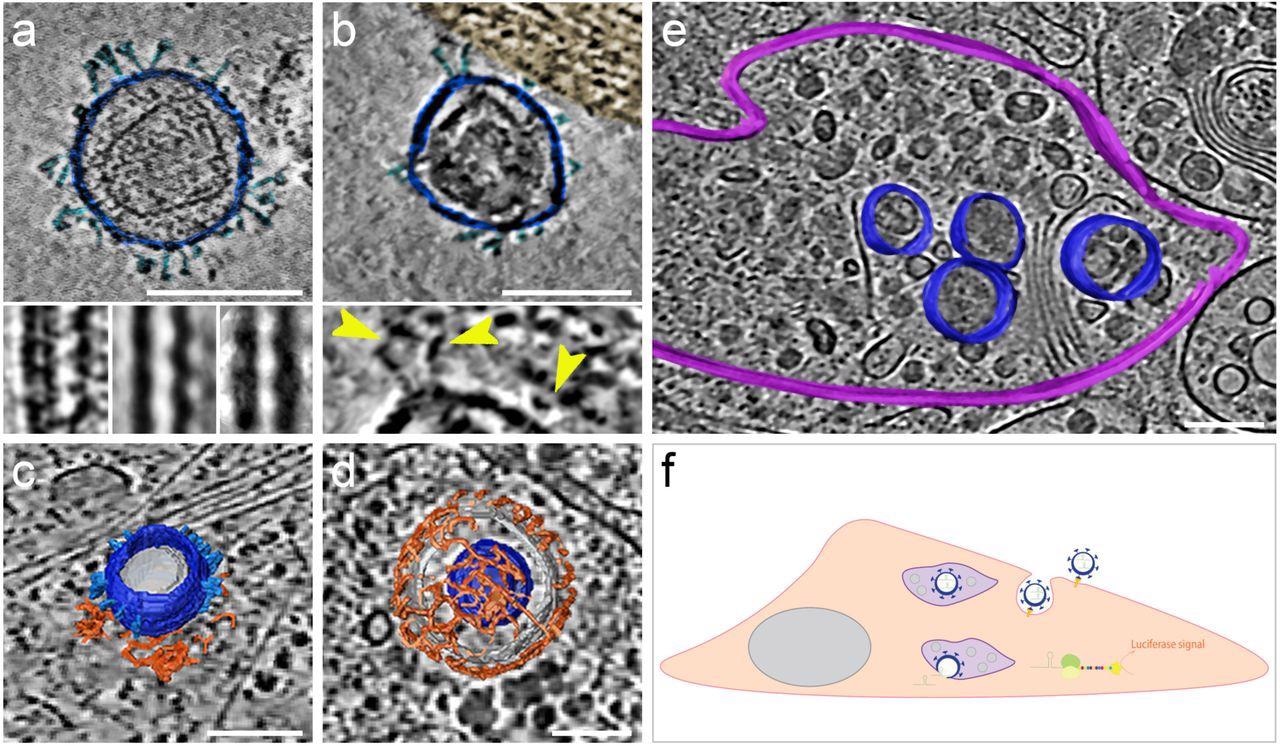
SARS-CoV-2 pseudoviruses enter cells via the clathrin-mediated endocytotic pathway. Cryo-ET reveals pseudovirus morphology and entry mechanism into Vero-E6 cells. Pseudovirus membranes and their segmentation are coloured dark blue. Spikes are coloured light blue. Clathrin triskeletons are coloured in orange, endolysosomal membrane in purple. All virtual slices are 12 nm thick. All scale bars = 100 nm. (a) Slice through a tomogram showing a pseudovirus. SARS-CoV-2 spikes are clearly identifiable. The pseudoviruses carry a distinct structural signature imposed by the MLV capsid lattice. The insets show the ridge like structure in a raw slice (left) and after averaging outside (centre) and inside (right) the cell. (b) Slice through a region of interaction between cell plasma membrane and a pseudovirus. The cell region, tinted gold, shows actin filaments running parallel to the plasma membrane. The inset shows a close-up view with yellow arrowheads pointing at connections between the membrane and the SARS-CoV-2 spikes of the pseudovirus. (c) Surface representation of a pseudovirus in the process of entering, attached to a clathrin-coated pit. A slice of the underlying tomogram is superimposed in the background for reference SARS-CoV-2. (d) Surface representation of a SARS-CoV-2 pseudovirus inside a clathrin-coated vesicle. A slice of the underlying tomogram is superimposed in the background for reference. (e) Surface representation of an endolysosomal compartment containing several pseudoviruses. A slice of the underlying tomogram is shown for reference and represents several loaded vesicles and convoluted membrane structures. (f) Schematic representation of SARS-CoV-2-pseudovirus entry and the subsequent steps leading to luciferase signals. Pseudoviruses bind hACE2 receptors via their SARS-CoV-2 spikes on the cell surface (b), which triggers endocytosis (c, d). Once inside, these pseudoviruses are targeted to the endolysosomal system (purple, e) allowing the release of luciferase mRNA.
The researchers found that hACE2 nanoparticles diminished the entry of pseudoviruses by 97.6% for MLVs with WT S protein, 97.4% for those with Delta S protein, and 99.4% for Omicron S protein pseudo-typed MLVs. Moreover, the team revealed that entry suppression using hACE2 nanoparticles was more efficient than using soluble hACE2. Further, they evaluated the effect of hACE2 nanoparticles on the replication kinetics of SARS-CoV-2 Delta using ex vivo explant cultures of human bronchus tissues. Observations revealed that viral replication was abolished at the limit of detection.
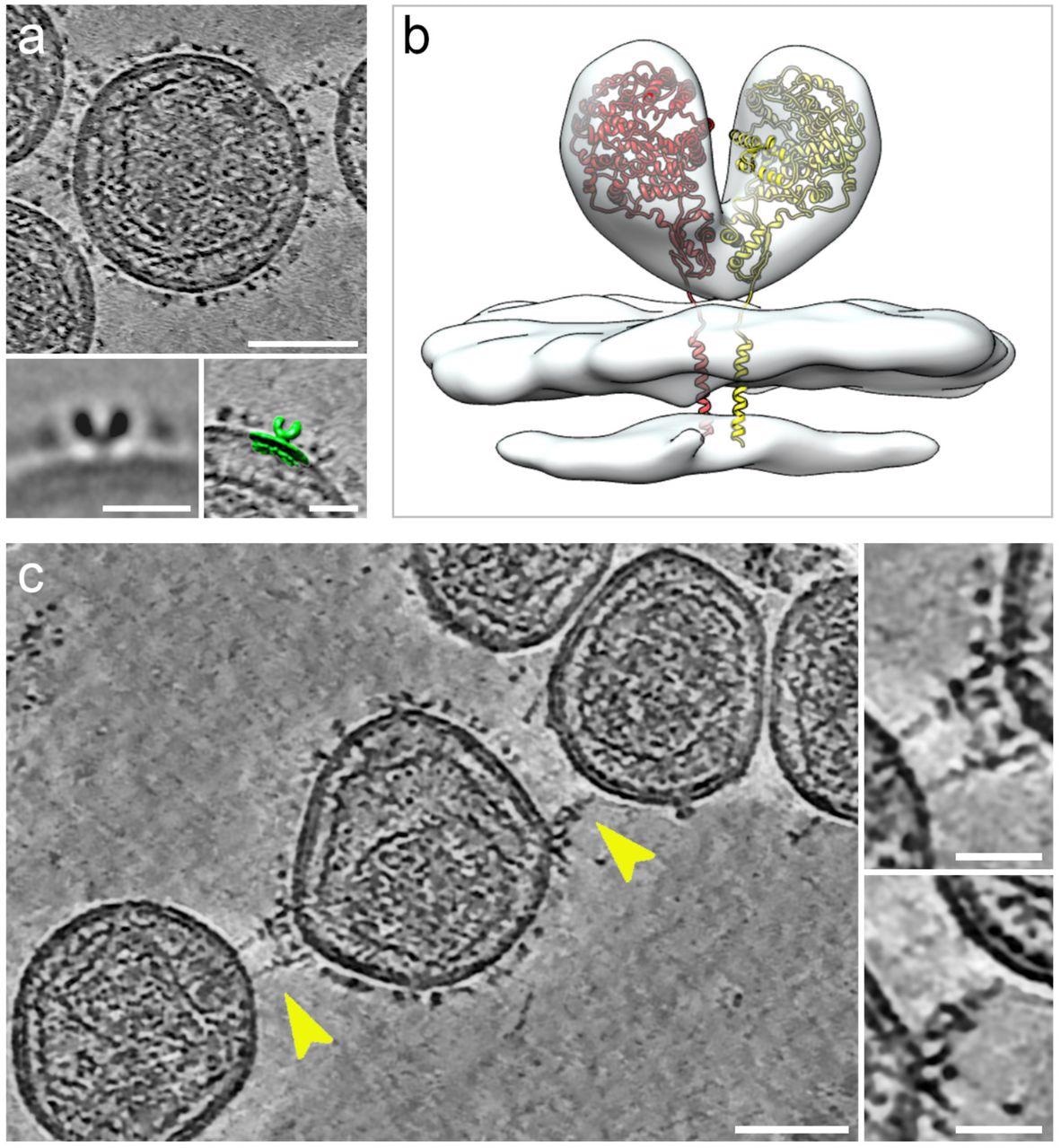
Cryogenic electron tomography of hACE2 nanoparticles reveals their interaction with SARS-CoV-2 pseudoviruses.All virtual slices in this figure are 12-nm thick. Bars in main panels = 50 nm; bars in insets = 20 nm. (a) Slice through a tomogram of an hACE2 nanoparticle showing dense decoration with hACE2 dimers. Insets show two-dimensional average of the protein density protruding from the nanoparticles (left) and overlay with surface representation (green) of three-dimensional (3D) reconstruction of membrane-bound particles (right). (b) Surface representation of 3D reconstruction obtained by sub-tomogram averaging of membrane-bound particles (Extended Data Figure 2). The atomic structure of the ACE2 dimer (monomers in yellow and red), extracted from the open conformation of the hACE2-B0AT1 complex (PDB: 6MD1), was fitted into the density showing a good match. (c) Virtual slice through a tomogram of hACE2 nanoparticles interacting with SARS-CoV-2 pseudoviruses. Interactions between hACE2 and spikes are marked with yellow arrowheads. Insets show close-up views of interactions between hACE2 nanoparticles and spike marked by the yellow arrowheads.
The interactions of hACE2 nanoparticles and pseudoviruses of SARS-CoV-2 were characterized using cryo-ET analysis. Structurally, the high-density particles protruding from the membrane of MLV were consistent with the membrane-embedded dimeric structures of ACE2. hACE2 nanoparticle and S-typed pseudo viral interactions were facilitated by the interactions between hACE2 dimers (of decoys) and pre-fusion S protein (of pseudoviruses).
Conclusions
The current findings showed that hACE2 nanoparticles efficiently preclude Vero-E6 cell infection by SARS-CoV-2 pseudoviruses. Furthermore, the infection of ex vivo cultures of bronchus tissues by live SARS-CoV-2 was prevented by hACE2 nanoparticles competing with cellular ACE2 and crosslinking ACE2 on the same nanoparticle engaged with multiple viruses.
Taken together, these results suggested that nanoparticles of hACE2 could act as efficient and potent decoys with robust neutralization of cell entry, representing a next generation of therapeutic interventions that are less susceptible to viral escape mechanisms than available strategies.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
ACE2 nanoparticles prevent cell entry of SARS-CoV-2. Cecile Sauvanet, Moara Lemos, Armel Bezault, Borja Rodriguez de Francisco, Michael CW Chan, Kenrie PY Hui, Ka-chun Ng, John M Nicholls, Niels Volkmann, Dorit Hanein. bioRxiv 2022, DOI: https://doi.org/10.1101/2022.05.05.490805, https://www.biorxiv.org/content/10.1101/2022.05.05.490805v1