Since its emergence in late 2019, SARS-CoV-2 has evolved substantially, with enhanced transmissibility and resistance to immunity. SARS-CoV-2 Omicron (BA.1) variant, first reported in November 2021, was soon designated as a variant of concern (VOC) by the World Health Organization (WHO) due to the alarmingly high number of mutations. The BA.1 variant quickly replaced predominant variants and wreaked havoc with an unprecedented surge in infections, including vaccine-breakthrough cases and reinfections.
Moreover, the growing frequency of novel subvariants of SARS-CoV-2 Omicron, such as BA.2.12.1, BA.4, and BA.5, further escalates the concerns about the evasion of immunity induced by vaccination or infection. The BA.2.12.1 variant, a descendent of Omicron BA.2, is spreading in the United States (US), whereas BA.4 and BA.5 variants have become dominant in South Africa. The BA.4 and BA.5 variants (hereafter BA.4/5) share identical spike (S) proteins, albeit have unique mutations relative to other subvariants.
 Study: Differential Evasion of Delta and Omicron Immunity and Enhanced Fusogenicity of SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. Image Credit: Juan Gaertner
Study: Differential Evasion of Delta and Omicron Immunity and Enhanced Fusogenicity of SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. Image Credit: Juan Gaertner

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
In the present study, researchers determined the neutralizing antibody (nAb) titers against SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 subvariants in sera from vaccinated or convalescent health care workers (HCWs). Additionally, they evaluated S proteins' infectivity, processing, and fusion properties from these Omicron sub-lineages. Finally, the infectivity of Omicron subvariants was tested using pseudo-typed lentivirus particles to infect CaLu-3 or HEK292T-ACE2 cells.
The authors noted that the infectivity of Omicron BA.1 was marginally higher than the D614G variant but approximately six-fold more heightened than the Delta variant in 293T-ACE2 cells, but BA.1 infectivity in CaLu-3 cells was 7.5-fold and 5.6-fold lower relative to D614G and Delta variants, respectively. BA.2 infectivity was comparable to BA.1, whereas other subvariants exhibited modest increases in infectivity. A 2.5- and a 3.8-fold increase was observed for BA.4/5 and BA.2.12.1 subvariants relative to D614G.
All Omicron subvariants showed lower infectivity in CaLu-3 cells than the D614G or Delta variant. nAb titers against Omicron subvariants were assessed using a pseudo-typed lentivirus neutralization assay. Fifteen HCWs vaccinated with two BNT162b2 or mRNA-1273 vaccine doses were evaluated for nAbs in their sera. The team noted a comparable resistance to neutralization by the Omicron subvariants with a 20-fold lower nAb titer relative to D614G.
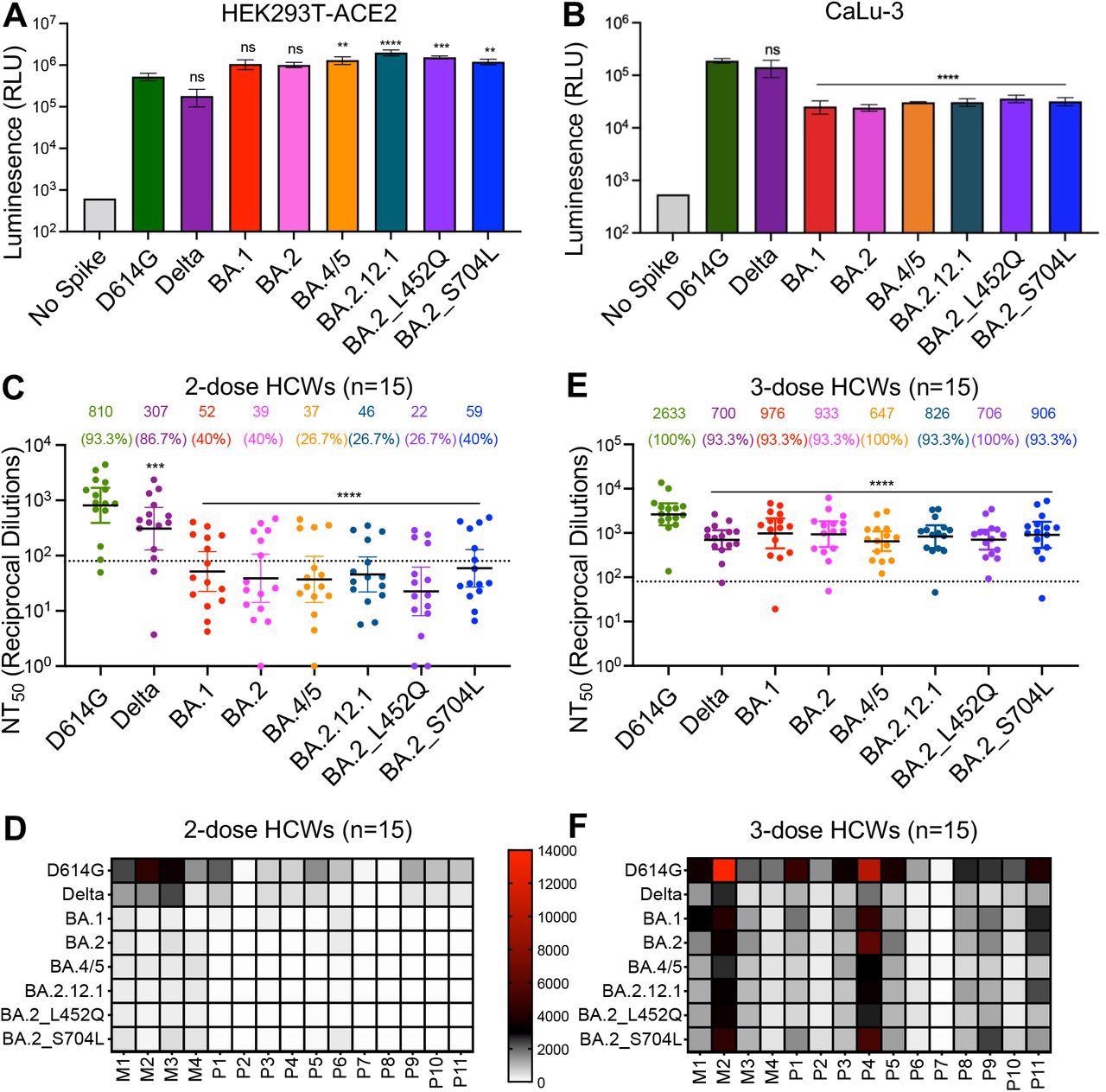
BA.4/5 and BA.2.12.1 subvariants exhibit stronger immune escape than BA.1 and BA.2. (A) Infectivity of pseudotyped viruses in HEK293T cells stably expressing ACE2 (HEK293T-ACE2). (B) Infectivity of pseudotyped lentivirus in human lung epithelia-derived CaLu-3 cells. Bars in (A) and (B) represent means ± standard deviation, and significance is determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction. Results of at least 3 independent experiments are averaged and shown. (C) Sera from 15 HCWs collected 3-4 weeks after second mRNA vaccine dose was used to neutralize pseudotyped virus, and the resulting geometric means of the 50% neutralization titers (NT50) are displayed at the top of the graph along with the percent of individuals with NT50 values above the limit of detection (NT50 = 80; dotted line). (D) A heat map showing patient/vaccinee NT50 values against each variant for the 2-dose HCW sera. (E) Sera from 15 HCWs following homologous mRNA booster vaccination were assessed for nAb titers. Bars in (C) and (E) represent geometric mean ± 95% confidence interval, and significance relative to D614G is determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction. (F) A heat map showing patient/vaccinee NT50 values against each variant for the 3-dose HCW sera. Patient/vaccinee numbers are identified as “P” for Pfizer/BioNTech BNT162b2 vaccinated/boosted HCW, “M” for Moderna mRNA-1273 vaccinated/boosted HCW. Throughout, p-values are represented as **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant.
Notably, the BA.2 variant with the L452Q substitution exhibited the highest resistance with a 36-fold lower nAb titer than the D614G strain. Sera from boosted individuals (third dose recipients) showed higher and improved nAb titers. Next, sera from infected individuals admitted to intensive care units (ICUs) during the Delta wave were examined. Sera from these patients showed high nAb titers against the Delta variant, albeit lower nAb titers were observed for BA.1 and BA.2 variants.
Interestingly, the BA.2.12.1 and BA.4/5 variants showed less escape from neutralization by the convalescent sera and had higher nAb titers. Those infected with BA.1 variant during the Omicron wave exhibited less potent neutralization of the emergent Omicron subvariants, particularly the BA.4/5 variant.
Nevertheless, a BNT162b2 or mRNA-1273 vaccine booster dose significantly enhanced nAb titers against all tested variants. The researchers studied membrane fusion by S proteins from different SARS-CoV-2 variants. The BA.1 variant showed a 4.7- and 11.3-fold lower fusogenicity than D614G and Delta variants. The BA.4/5 and BA.2.12.1 variants exhibited an increased propensity of fusion than either BA.1 or BA.2 variant, although much lower than for the Delta variant.
The expression of S protein on the surface of virus-producing cells was analyzed using flow cytometry. BA.1 or BA.2 S protein expression was slightly higher than D614G or the Delta variant. The emergent Omicron subvariants had similar surface expression. Last, the team evaluated the processing of S protein in lysates of virus-producing cells and purified virus particles.
Previously, the authors reported a lower propensity of BA.1 S protein for furin cleavage, indicated by the lower S1/S ratio. The processing of BA.1 and BA.2 S protein was comparable, with slightly enhanced processing of BA.2.12.2 and BA.4/5 S proteins with approximately 1.4- and 1.2-fold increase relative to D614G S protein. Similar results were observed for purified viral particles, and it has been speculated due to the L452Q/R substitution.
Conclusions
The authors analyzed the immune characteristics induced by vaccines or natural infection against the SARS-CoV-2 Omicron BA.2.12.1 and BA.4/5 subvariants, including their S protein fusogenicity and furin cleavage features. There was no evidence that two doses of either vaccine were adequate to neutralize SARS-CoV-2 Omicron variants.
Although booster doses amplified neutralizing titers, they were less potent against the emergent Omicron subvariants. The researchers revealed that substitutions on the residue 452 of S protein might be critical and could drive resistance to neutralization. Overall, these findings emphasize the need for continuous surveillance of new variants as well as a detailed understanding of S protein biology and its influence on SARS-CoV-2 pathogenicity and immunity.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Differential Evasion of Delta and Omicron Immunity and Enhanced Fusogenicity of SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants, Panke Qu, John P Evans, Julia N. Faraone, Xue Zou, Yi-Min Zheng, Claire Carlin, Joseph S. Bednash, Gerard Lozanski, Rama K. Mallampalli, Linda J. Saif, Eugene M. Oltz, Peter J. Mohler, Richard J. Gumina, Shan-Lu Liu, bioRxiv 2022, DOI: https://doi.org/10.1101/2022.05.16.492158, https://www.biorxiv.org/content/10.1101/2022.05.16.492158v1