Recent mutational changes in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein, particularly within the receptor-binding domain (RBD) and receptor binding motif (RBM), have allowed its immunogenic escape from vaccination-elicited immunity. In a recent study published on the preprint server bioRxiv,* researchers utilize the ATLAS screening assay to reveal the antigen response profiles of T-cells to different SARS-CoV-2 variants.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
Peripheral blood mononuclear cells (PBMCs) were collected from three cohorts of donors including unexposed individuals, as well as those who experienced severe and mild illness following SARS-CoV-2 infection. As no difference could be found between the severe and mild cohorts, these were later collated together.
All exposed subjects were required to have their SARS-CoV-2 infection confirmed through reverse transcription-polymerase chain reaction (RT-PCR) assay. The ATLAS assay was then used to identify CD4+ and CD8+ T-cells by utilizing the endogenous human leukocyte antigen (HLA) class I and II peptide processing pathways.
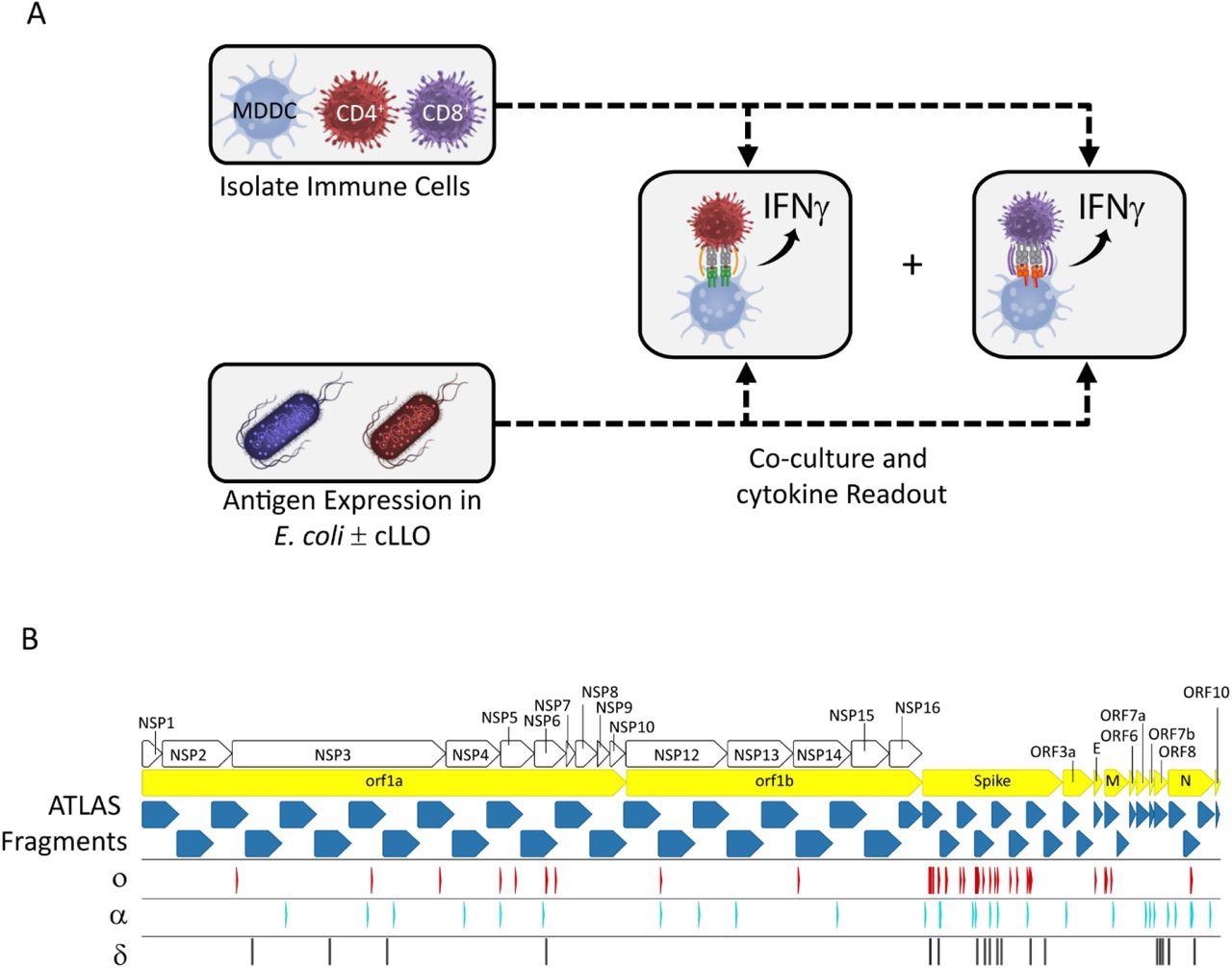
The ATLAS assay and library design. (A) Each putative antigen is expressed individually in two E. coli strains one with and one without cLLO (bottom left box), which are arrayed in expression libraries. Monocyte-derived dendritic cells (MDDCs) as well as CD4+ and CD8+ T cells are isolated from peripheral blood and co-cultured overnight with the E. coli libraries and IFNγ output measured in each well. (B) Schematic representation of the library design depicting the location of the ATLAS Fragments (blue chevrons) in relation to the SARS-CoV-2 orfs (yellow chevrons) and orf1ab NSPs (white chevrons). Mutations from the Omicron (o), Delta (δ), and Alpha (α) VOCs are shown as red, cyan, and black barcodes, respectively.
Study findings
Both exposed and unexposed cohorts were profiled using the ATLAS library, which revealed a wide range of responses across the SARS-CoV-2 proteome, with every fragment producing either a CD4+ response, CD8+ response, or both. In all SARS-CoV-2 open reading frames (ORFs) other than ORF7b, to which no donor showed a CD8+ response, both T-cell types showed responses to ATLAS fragments.
T-cell responses appeared to favor the spike, membrane, and nucleocapsid proteins, as the number of responses per fragment for ORFs was larger than the rest of the proteome. Other biases could be observed when the response profiles of T-cell subsets were compared, with several ORFs and the C-terminus of the nucleocapsid protein almost exclusively targeting CD4+ T-cells.
Biases were also observed for ORF1ab, with CD4+ responses localized to a few fragments in ORF1a. However, these biases spread throughout ORF1b, with CD8+ responses following the opposite pattern.
Common and frequent antigens for both T-cell types were identified in both cohorts, particularly in ORF1ab, as well as the nucleocapsid and spike proteins. Comparatively, the membrane protein and ORF10 only encoded common CD4+ antigens.
Unexposed donor samples showed responses to multiple SARS-CoV-2 antigens, with all donors showing at least one response. Moreover, 72% and 90% of fragments induced at least one CD4+ or CD8+ response, respectively, with a comparison of unexposed and exposed profiles revealing that both cohorts responded to similar regions of the proteome at comparable frequencies.
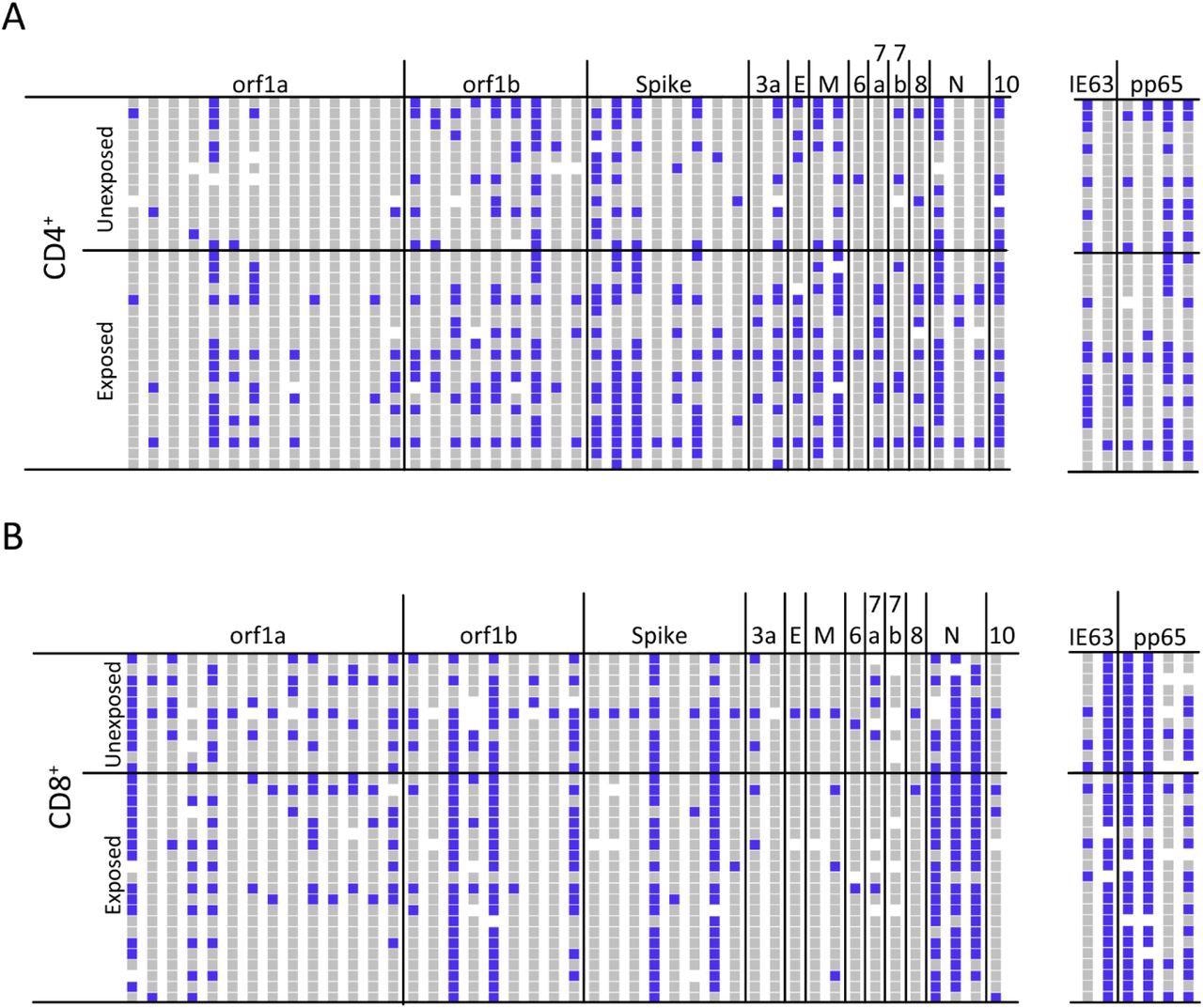
ATLAS identifies common and frequent SARS-CoV-2 antigens. (A) The CD4+ T cell response profiles to SARS-CoV-2 (left) and control antigens (right) for unexposed (N = 14) and exposed (N = 20) donors. Responses are blue, non-responses are grey, and white indicates not tested. Each column represents an ATLAS clone, and each row is a donor. Fragments and donors are grouped by orf and cohort, respectively. (B) The CD8+ response for unexposed (N = 11) and exposed (N = 21) donors are reported similarly.
The similarities between both cohorts resulted in the inability to perform statistical analyses to examine the difference between those who had been exposed and those who had not. Some unexposed donors in previous studies have shown responses to SARS-CoV-2 antigens, which is likely due to exposure to other coronaviruses. Notably, the ATLAS assay can be configured to identify infrequent cross-reactive antigens and avoid this issue.
Spike peptide fragments were designed so that S_2-178 and S_159 encoded the N terminal domain (NTD), S_316-492, and S_473-649 span the RBD/RBM, and the remaining four fragments were spread across the C-terminus.
The SARS-CoV-2 Alpha and Delta mutations were mapped onto these fragments, which revealed that polymorphisms were mostly present in the first half of the NTD and central portion of the protein. Both variants showed no more than three mutations in any one fragment.
Comparatively, SARS-CoV-2 Omicron mutations outnumbered those of the other variants in every fragment other than the last, which had none. A total of 12 Omicron mutations were mapped to fragments that encoded the RBD, with the highest density recorded in the region most critical for angiotensin-converting enzyme 2 (ACE2) binding.
Similar to the results for the rest of the genome, the unexposed and exposed recognition profiles showed significant overlaps. A complementary pattern could be observed when comparing T-cell subsets, with CD8+ T-cells from nearly all donors recognizing at least one spike fragment and CD8+ cells from 75% of donors recognizing at least two. Similar frequencies were reported for CD4+ T-cell responses.
Most of the CD8+ responses were localized to two fragments, which were identified as frequent antigens, one of which encodes for the RBM. CD4+ T-cell responses were found across the length of the protein, although few class II responses could be identified at the C-terminal end.
Conclusions
The ATLAS assay can maximize the ability of the immunoproteasome to generate and present optimally processed peptides. In the current study, T-cell responses were detected throughout the SARS-CoV-2 proteome, with several regions successfully characterized as common or frequent antigens to corroborate previously reported data.
Taken together, these findings could help better understand the impact of SARS-CoV-2 Omicron mutations, as well as those present in other SARS-CoV-2 variants.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.