Single-cell RNA-sequencing (scRNA-seq) shows which genes are turned on in a cell and what their level of transcription is. This allows in-depth assessment of the biology of individual cells, and detection of changes that may indicate disease. scRNA-seq is becoming widely used across disciplines including developmental biology, neurology, oncology, immunology, cardiovascular research and infectious diseases. It is critical for studying population heterogeneity, identifying minority sub-populations of interest, and for discovering unique characteristics of individual cells. Applications reach far beyond ophthalmology: single-cell sequencing is widely used for the analysis of de novo germline mutations and somatic mutations in normal and diseased cells, e.g. in cancer cells.
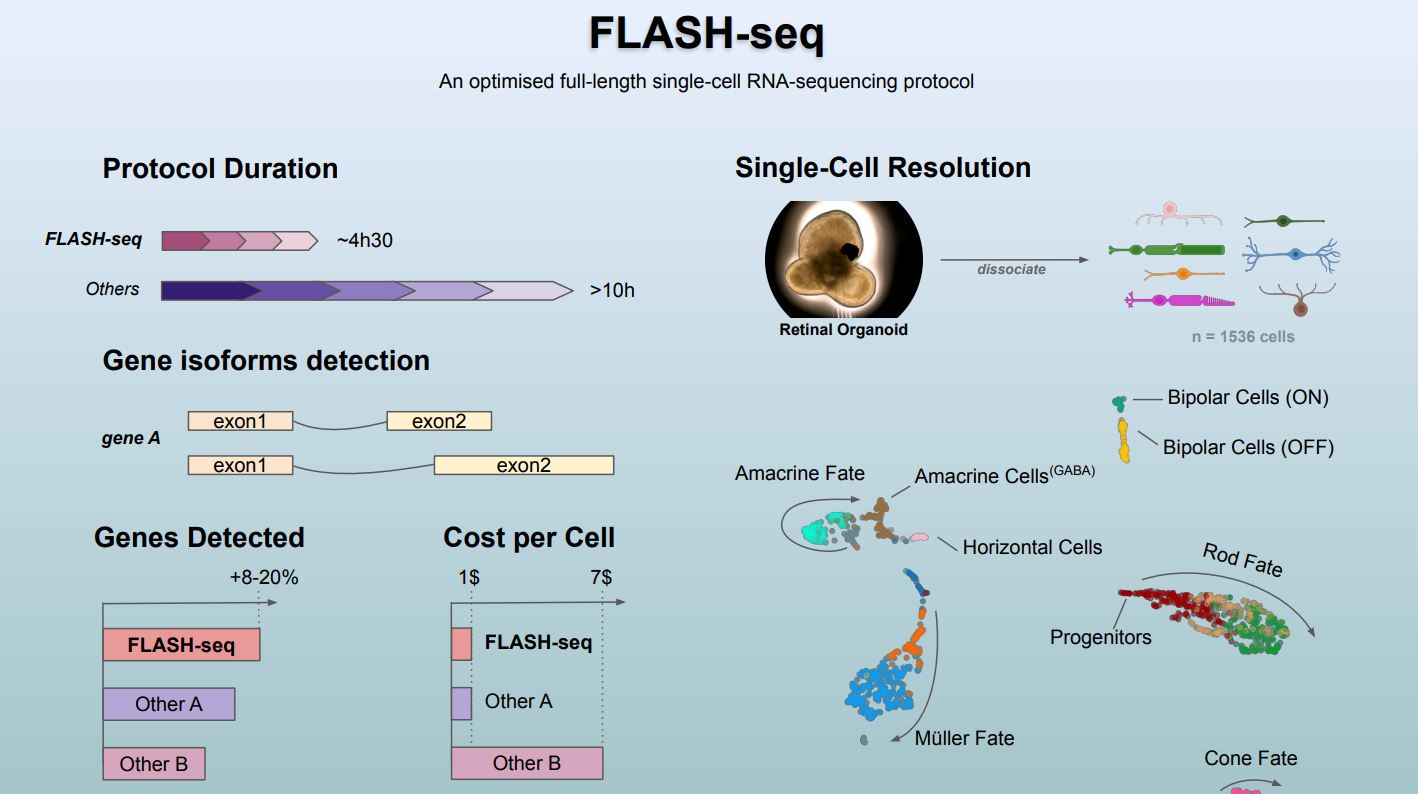
FLASH-seq: An optimized full-length single-cell RNA-sequencing protocol. Image Credit: Institute of Molecular and Clinical Ophthalmology Basel
A new scRNA-seq protocol developed at IOB, with scientists from the Novartis Institutes for BioMedical Research, can be used to study any disease model requiring the analysis of rare cell populations at high resolution.
Our modular FLASH-seq protocol provides a snapshot of the cell transcriptome at an unprecedented resolution. The method can be miniaturized, automated and adapted to different needs. It helps to define which gene isoforms are present in health and disease. It also provides a much deeper picture of the gene expression, especially after perturbation due to disease, developmental defects or external agents. Moreover, it is easy to set up in the lab, 50% faster and cheaper than similar existing protocols and enables the study of molecular mechanisms of disease beyond the scope of current single-cell sequencing tools.”
Simone Picelli, Head of IOB Single-Cell Genomics Platform and Senior Author of Paper
The new method can generate sequencing-ready libraries in just half a day. Researchers at IOB therefore believe that FLASH-seq has the potential to become the tool of choice when looking for an efficient, robust, modular, affordable and automation-friendly full-length scRNA-seq protocol.
Source:
Journal reference:
Hahaut, V., et al. (2022) Fast and highly sensitive full-length single-cell RNA sequencing using FLASH-seq. Nature Biotechnology. doi.org/10.1038/s41587-022-01312-3.