The spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has waxed and waned, depending on the level of population immunity and the biological characteristics of the virus. The emergence of new variants of the virus, with higher transmissibility or immune escape characteristics, for instance, has changed the course of the pandemic. A new research paper in the journal Viruses reports the isolation of a recombinant strain in a patient infected with the virus for over a year.
Genetic recombination refers to the combination of corresponding parts of a genomic molecule to form a new sequence. It commonly occurs in ribonucleic acid (RNA) viruses, including the currently circulating SARS-CoV-2, a member of the family of coronaviruses. In fact, it is suspected to have given rise to the pandemic virus.
This phenomenon has also been inferred to occur in human patients with this infection, as shown by the presence of the distinctive mutations of different strains of the virus in the same patient. However, none of these has been isolated so far.
In the current paper, published in the journal Viruses, the scientists found evidence of the slow emergence of a recombinant strain of SARS-CoV-2 in a chronically immunocompromised patient. The patient was a 56 years old male with lymphoma.
The first positive test came in August 2020, when he developed pneumonia, requiring intensive care unit (ICU) admission. This was successfully treated, but the virus was not cleared. During the last 14 months of his life, he repeatedly tested positive for the virus before dying of complications following the lymphoma and progressive multifocal leukoencephalitis first diagnosed in May 2021.
The first detection of a hybrid-genome SARS-CoV-2 variant came after 14 months of infection. The two strains that had undergone recombination were identified as B.1.160, and the Alpha variant. Two regions of the former had been replaced by the latter, and the spike gene showed all the eight characteristic mutations of the Alpha variant, while the S477N mutation which is a hallmark of B.1.160 was absent.
There was little evidence to suggest that the presence of these Alpha mutations was due to coinfection or contamination by both variants. Instead, “These findings indicated that this mosaic genome was the result of recombinations between parental genomes of the B.1.160 and Alpha variants.”
The timeline of positive viral RNA tests suggested that the initial infection occurred with B.1.160, circulating widely from August 2020 until January 2021, when the Alpha variant replaced it. The next sequence was obtained only 8 months later, and this showed that a mosaic genome had already been formed between the two strains.
This recombinant formed over several steps, with recombination occurring at three sites of the two parental genomes. The detection of multiple chimeric copies indicated the presence of several breakpoints between the ancestral strains.
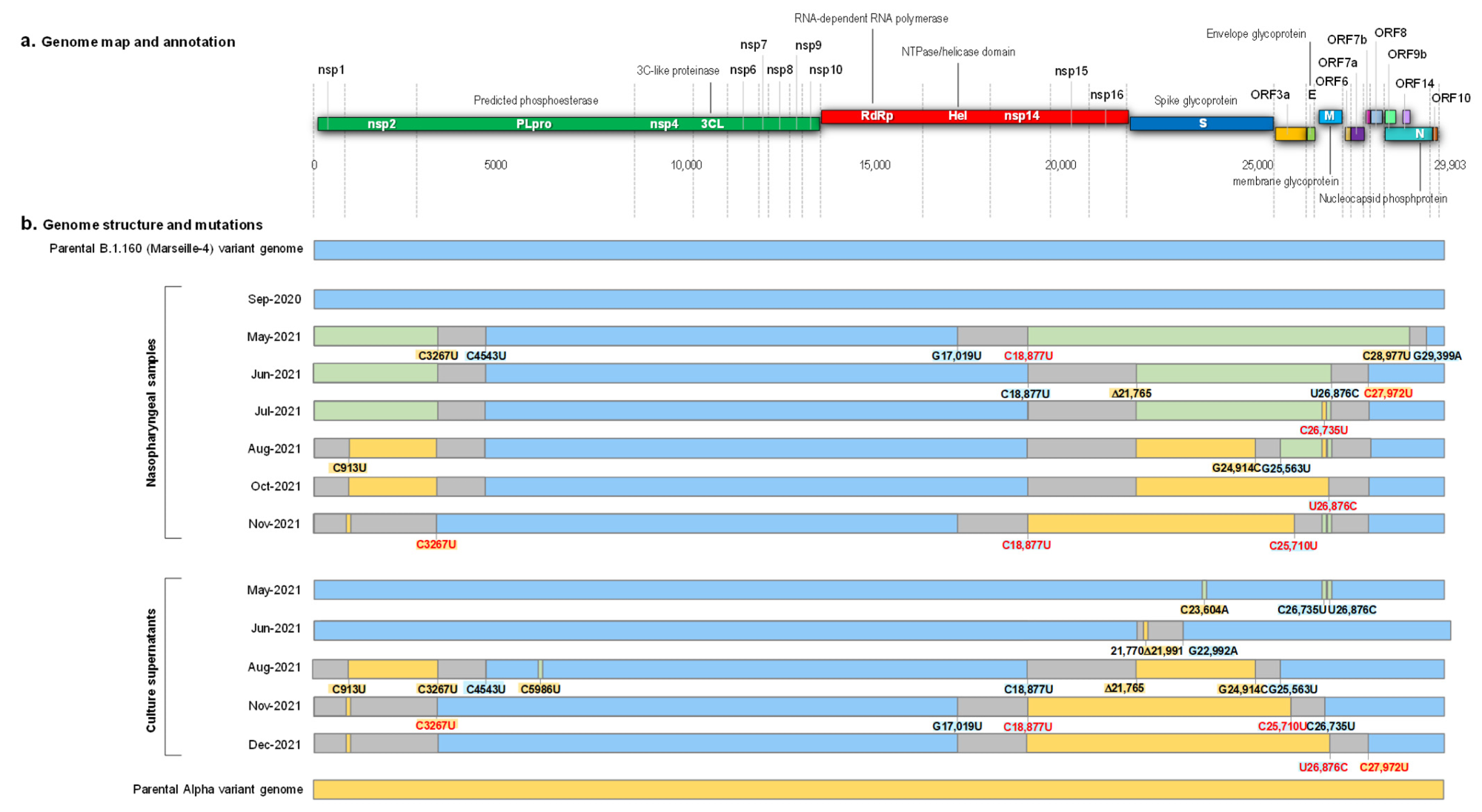
Schematic representation of the structure of the SARS-CoV-2 genomes obtained from the nasopharyngeal samples and from the culture supernatants, as well as of the recombination events over time, in reference to parental genomes of the B.1.160 and Alpha variants. (a) Genome map and annotation; (b) Genome structure and mutations. Blue color of rectangles indicates sequences from a B.1.160 variant; yellow color indicates sequences from an Alpha variant; green color indicates co-detection of sequences from a B.1.160 variant and from an Alpha variant; grey color indicates sequences from indeterminate origin. Signature mutations from the B.1.160 and Alpha variants are indicated by a blue background and a yellow background, respectively. Signature mutations that are absent are indicated by a red font. Δ21,765: -6 nucleotides; Δ21,991: -3 nucleotides. Nsp, nonstructural protein; ORF, open reading frame.
What Are the Implications?
The results of this study emphasized the presence of recombination in an immunocompromised lymphoma patient who developed a prolonged course of infection with SARS-CoV-2 despite undergoing multiple treatments. The outcome was the development of a hybrid virus strain formed by B.1.160 and the Alpha variant. The replacement of a distinctive B.1.160 mutation by several characteristic Alpha mutations between 8 and 14 months were thought to be non-accidental, and the result of recombination at three sites.
Further analysis of the genomic samples from successive respiratory samples and cultures of the virus showed a stepwise development of several hybrid viruses in this patient, among which one established itself and continued to replicate in the host until the very end.
This early sampling study, which covered over a year, yields much insight into how RNA viruses recombine to form new strains, and in particular, reveals the generation of a recombinant SARS-CoV-2 strain that was isolated in culture. Earlier research has shown the presence of 16 Alpha-non-Alpha recombinants in 2021 in the UK. Others have suggested that recombinants account for 5% of circulating viruses in the UK and USA.
Even as more recombinants are being detected every month, researchers are exploring different pathways in various cell types or between different types of RNA viruses. This could help understand how human pathogenic RNA viruses emerge.
The significance of such findings should “lead to a strengthening of genomic surveillance in patients, especially in immunocompromised long-term viral carriers. Such infectious episodes could perhaps lead to the emergence of new emerging viruses.”