The US Centers for Disease Control and Prevention (CDC) tracking project pointed at several regional differences in the COVID-19 incidence, fatalities, and health disparities. Therefore, it became crucial to curate, integrate, and analyze COVID-19 data at the regional levels and aggregate the results to inform national-level policies.
A data commons, such as PRC, curate, integrate, and harmonize data for a specific community, e.g., researchers studying an epidemic or pandemic, public health workers, and policymakers. Typically they require several legal and data agreements.
However, a regional instance of a PRC developed in the current study was designed to be part of a broader data ecosystem, operate at a low level, and increase activity as required by the pandemic. Most importantly, it comprised multiple regional commons to support the pandemic response through local, regional, and federated data sharing and analysis.
About the study
In the present study, researchers used the open-source Gen3 data platform to develop PRC, and a formal consortium of Chicagoland area organizations operated it. Gen3, based upon consortium, data, and platform agreements, was developed by the non-profit Open Commons Consortium.
The Open Commons Consortium has three main functions, as follows:
i) it helps establishment of a consortium to build and operate a data commons,
ii) ensures data is contributed to a data commons, and
iii) facilitates its members to work in groups, analyze data, and develop software applications and services to enhance the functionality of the commons.
The CCC curated and harmonized several datasets, including clinical data of ~90,000 patients, statistical data summary of COVID-19 cases, and sequencing data of over 5,300 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant genomes.
Study findings
The CCC had eight members and five working groups. Its eight members, viz., Rush University Medical Center, University of Chicago, Southern Illinois University, the University of Illinois at Chicago, St. Anthony Hospital, Sinai Chicago, NorthShore University HealthSystem, and CommunityHealth, had contributed clinical data from over 90,000 subjects with COVID-19.
The clinical data working group developed a standard data model for each member to contribute data in the required format. The epidemiological modeling working group used the CCC-obtained aggregated counts for COVID-19 cases, deaths, and select comorbidities to understand health disparities and build predictive models. They developed hierarchical Bayesian models that predicted county-wise future COVID cases and fatality counts for Illinois. Likewise, the working group developed regression models to understand temporal, age-related race/ethnicity differences in case/fatality ratios. The variant surveillance working group collected and contributed over 5,300 SARS-CoV-2 genome sequences to national and international genomic databases.
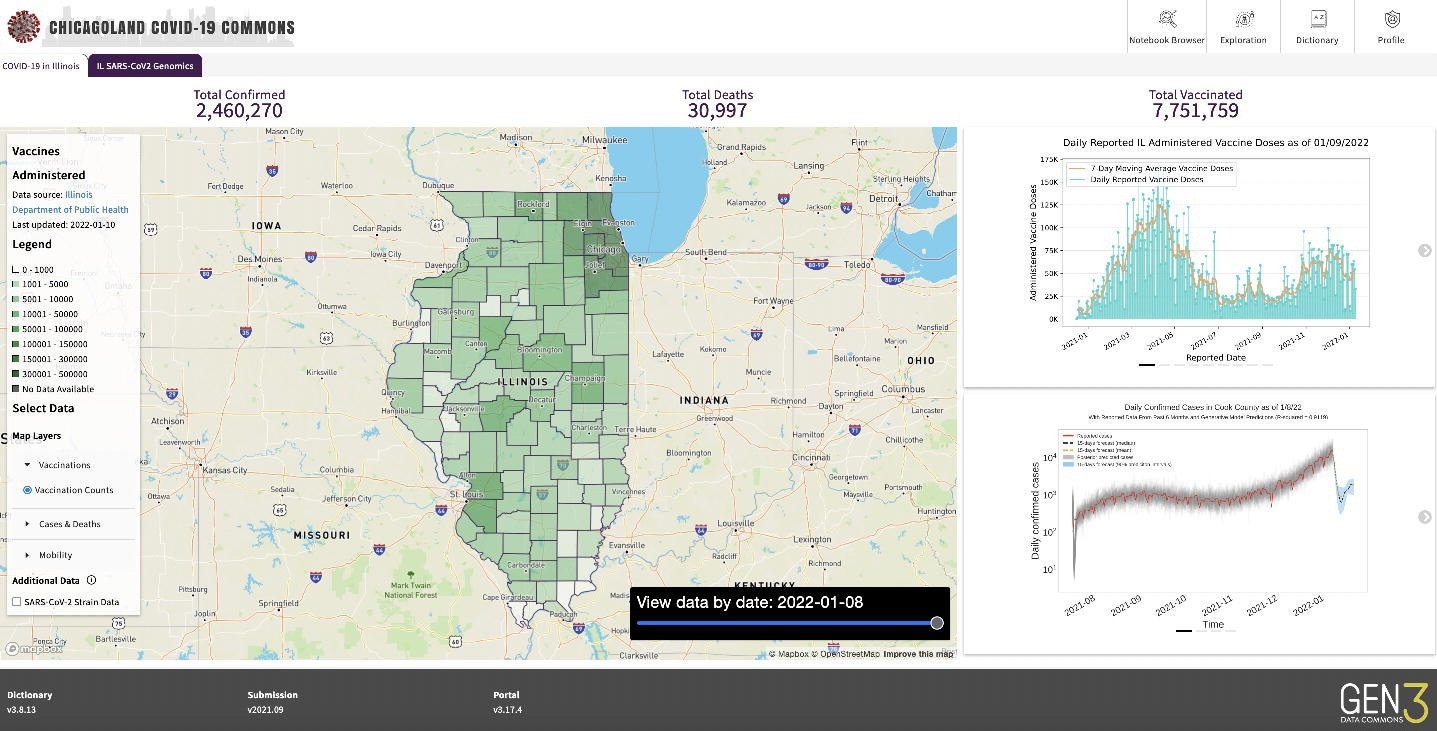 Screenshot of PRC
Screenshot of PRC
Gen3 software automatically generates application programming interface (APIs) for data and metadata access, data submission, authorization, and authentication, all of which make both controlled and public access findable, accessible, interoperable, and reusable (FAIR). For instance, PRC hosts a publicly accessible PRC Jupyter Notebook Browser that helps access COVID-19 case incidence, fatality, clinical, mobility, and imaging data.
Three participating institutions contributed patient-level COVID-19 data starting March 1, 2020. The PRC analyzed submitted data and identified data quality issues. The quality analysis included developing plots to compare patient counts by demography, symptoms, hospitalization events, and pre-existing comorbidities. Further, the PRC used statistical summary reports (SSR) county-level data to develop epidemiological models, which provided information for map overlays that are easily accessible to the public.
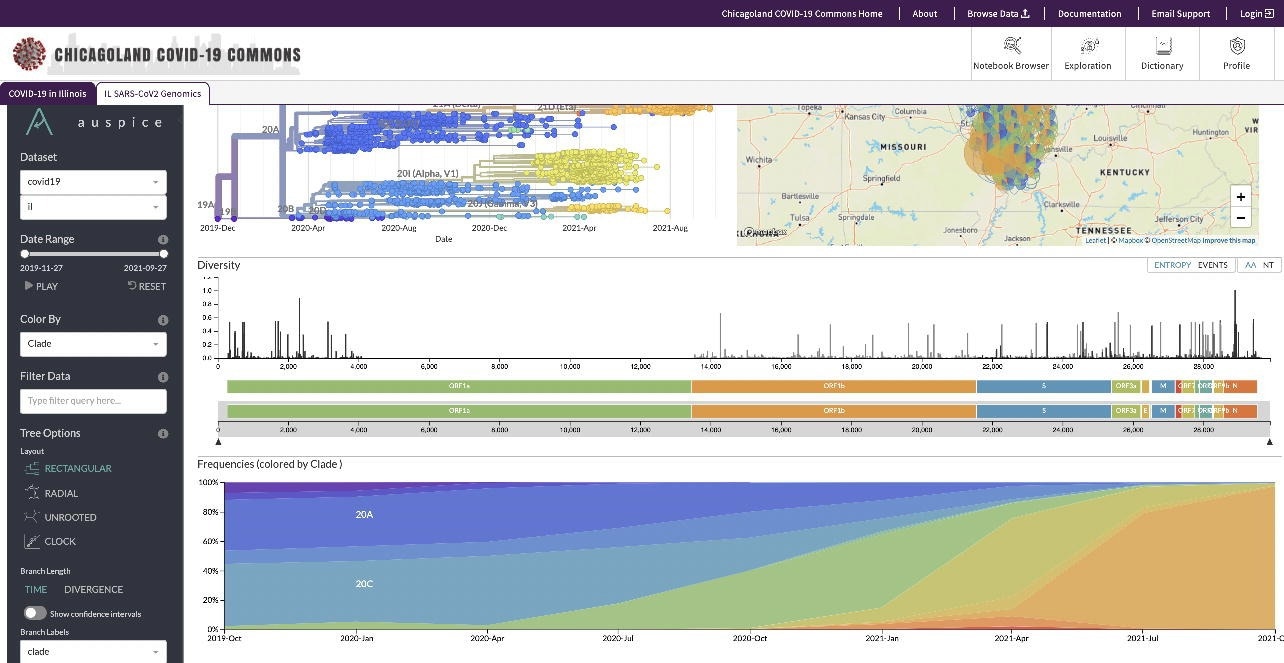
Screenshot of viral variants and their geographic distribution
The PRC also worked on a project with Southern Illinois University (SIU) to analyze the genomic sequence of SARS-CoV-2 and better understand the spread of COVID-19 across Illinois. The project had sequenced over 5,300 SARS-CoV-2 genomes spanning 16 viral clades and more than 150 variants to track SARS-CoV-2 evolution in Illinois and identify the appearance of specific SARS-CoV-2 variants of concern (VOCs).
Conclusions
The CCC contained clinical data from over 90,000 COVID-19, SSRs for the analysis of COVID-19 health disparities, over 5,300 SARS-CoV-2 genome sequencing data, and COVID-19-related public data. Overall, the CCC data was rich, readily available to a broader community, and enhanced the national view of COVID-19-related issues to accelerate research on COVID-19 and Long COVID. In summary, the study highlighted the significance of a regional COVID-19 commons in complementing the ongoing efforts to gather COVID-19 data at the national level to help support clinical research and policy development.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
The Pandemic Response Commons, Matthew Trunnell, Casey Frankenberger, Bala Hota, Troy Hughes, Plamen Martinov, Urmila Ravichandran, Nirav S Shah, Robert L Grossman, medRxiv pre-print 2022, DOI: 10.1101/2022.06.20.22276542v1, https://www.medrxiv.org/content/10.1101/2022.06.20.22276542v1