Scientists have successfully developed vaccines to protect individuals from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19) pandemic. However, all currently available COVID-19 vaccines have been designed based on the ancestral SARS-CoV-2 strain that emerged in 2019.
Background
Previous studies have shown that COVID-19 vaccines elicit an immune response that reduces disease severity and the incidence of infection. Additionally, vaccination has decreased the hospitalization rate due to COVID-19.
Since 2019, several SARS-CoV-2 variants have emerged due to continual genomic mutations. These variants have been categorized as variants of concern (VOC) and variants of interest (VOI) in accordance with their transmission rate, virulence, and capacity to evade immune protection induced through either vaccination or natural infection.
Currently, the SARS-CoV-2 Omicron VOC is the dominant circulating strain throughout the world and has been found to be resistant to neutralizing antibody (nAb) responses elicited after COVID-19 vaccination.
Reduced effectiveness of most approved therapeutic monoclonal antibodies and their mixtures, such as BRII-196, LY-CoV555, REGN10987, REGN10933, and LY-CoV016, have also been reported against Omicron infections. Thus, there is an urgent need for effective vaccines and/or therapeutics that could offer a broader range of protection against present and future SARS-CoV-2 VOCs and VOIs.
Typically, neutralizing potency and breadth of antibodies are considered to be mutually exclusive. The majority of current nAbs used to treat COVID-19 target the angiotensin-converting enzyme 2 (ACE2) receptor binding site (RBS) of the SARS-CoV-2 spike (S) protein; thus, because SARS-CoV-2 variants typically contain most of their mutations in the RBS region, nAbs are less effective.
One prior study has shown that nAbs like CR3022 that can target other epitopes on the receptor-binding domain (RBD) also exhibit reduced neutralization potency against SARS-CoV-2 variants.
About the study
A new Biophysics and Computational Biology study successfully identified a potent neutralization epitope in SARS-related coronaviruses (CoVs) including SARS-CoV-1, WIV1, and SHC014. In this study, scientists identified a region on the RBD of the SARS-CoV-2 S protein, which is targeted by a diverse group of antibodies. Moreover, the therapeutic antibody ADG20 was found to neutralize SARS-CoV-2 VOCs, such as the Omicron variant, as well as other CoVs.
One previous study showed that neutralization by ADG20 against the Omicron variant was reduced by 20-fold as compared to the Delta variant. This antibody was determined to be the most effective among several therapeutic antibodies.
The Omicron (BA.1) variant has evolved to produce the BA.2 (B.1.1.529.2) sublineage. As compared to the Omicron BA.1 sublineage, BA.2 contains one reversion (S446G) and three additional mutations of T376A, D405N, and R408S, along with other mutations such as S371F instead of S371L in its RBD.
Recent studies have shown that BA.2 escapes ADG20 and other antibodies like S2X259 targeting this site. This might be due to differences in the interaction between residue 371 and the N343 glycan.
Many therapeutic antibodies have been developed against SARS-CoV-2, some of which include sotrovimab, REGEN-COV, Evusheld, and bebtelovimab. Among these antibodies, bebtelovimab was found to be most effective against both Omicron BA.1 and BA.2. However, Evusheld was found to be effective against BA.2 but not BA.1.
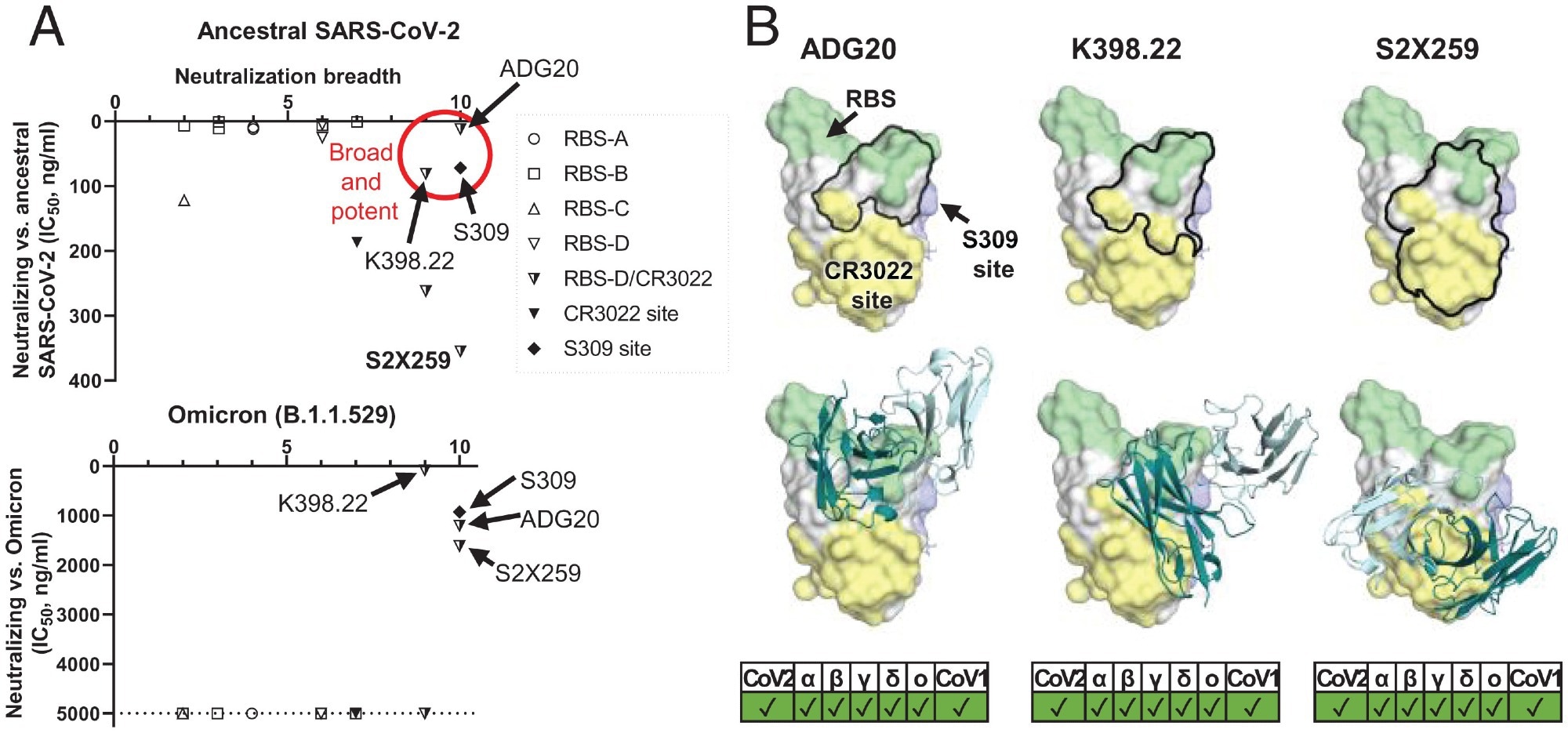 A common region on the RBD targeted by potent-and-broad nAbs against SARS-CoV-2. (A) A scatter plot of antibody neutralization potency (IC50, ng/mL) against (Top) ancestral SARS-CoV-2 and (Bottom) Omicron vs. neutralization breadth, defined by the number of SARS-related CoV strains neutralized in this study (10 strains in total, including ancestral SARS-CoV-2, Alpha, Beta, Delta, Gamma, Mu, Omicron, SARS-CoV-1, pang17, and WIV1). (B) All of the potent-and-broad nAbs target a region that spans from one end to the RBS (green) to the proximal CR3022 site (yellow). Epitopes of each antibody are outlined by black lines (Top), where the variable domains of ADG20, K398.22, and S2X259 are shown in cartoon representation with heavy chains in deep teal and light chains in light cyan (Bottom). Epitopes and the RBS are defined by residues with BSA of >0 Å2 as calculated by PISA (50) using structures of ADG20 (this study), K398.22 (26), S2X259 (PDB 7RAL) (57), CR3022 (PDB 6W41) (9), S309 (PDB 6WPS) (24), and ACE2 (PDB 6M0J) (54). SARS-CoV-2 variants as well as SARS-CoV-1 can be neutralized by these antibodies, as shown at the Bottom of the panel.
A common region on the RBD targeted by potent-and-broad nAbs against SARS-CoV-2. (A) A scatter plot of antibody neutralization potency (IC50, ng/mL) against (Top) ancestral SARS-CoV-2 and (Bottom) Omicron vs. neutralization breadth, defined by the number of SARS-related CoV strains neutralized in this study (10 strains in total, including ancestral SARS-CoV-2, Alpha, Beta, Delta, Gamma, Mu, Omicron, SARS-CoV-1, pang17, and WIV1). (B) All of the potent-and-broad nAbs target a region that spans from one end to the RBS (green) to the proximal CR3022 site (yellow). Epitopes of each antibody are outlined by black lines (Top), where the variable domains of ADG20, K398.22, and S2X259 are shown in cartoon representation with heavy chains in deep teal and light chains in light cyan (Bottom). Epitopes and the RBS are defined by residues with BSA of >0 Å2 as calculated by PISA (50) using structures of ADG20 (this study), K398.22 (26), S2X259 (PDB 7RAL) (57), CR3022 (PDB 6W41) (9), S309 (PDB 6WPS) (24), and ACE2 (PDB 6M0J) (54). SARS-CoV-2 variants as well as SARS-CoV-1 can be neutralized by these antibodies, as shown at the Bottom of the panel.
Study findings
The current study reports several mutations associated with the Omicron variant that reduced the neutralizing capacity of existing antibodies. For instance, LY-CoV016 binds to RBS-A of several SARS-CoV-2 variants to promote neutralizing activity. However, in the case of the Omicron variant, neutralization does not occur, as this variant contains mutation K417N, which results in the loss of a salt bridge.
Similarly, previous studies reported that antibodies AZD8895, REGN10933, and LY-CoV555 bind to the RBS-B of SARS-CoV-2 to promote neutralization. However, the presence of certain mutations like E484A and Q493R in the Omicron variant has led to a loss of polar interactions, which has reduced the neutralization potency of these antibodies. Additionally, the N440K mutation of the Omicron variant has reduced the binding potential of AZD1061, REGN10987, and LY-CoV1404 to the RBS-D site.
The S371F mutation in the BA.2 sublineage reduces the neutralizing capacity of sotrovimab. Hence, this antibody is effective against BA.1.
The results of the current study are consistent with structural observations of previous studies that have reported reduced or abolished neutralization of AZD8895 against BA.2.12.1, BA.4, and BA.5.
A class of antibodies that consist of long CDRs H3 with a YYDRxG motif have been discovered to target the highly conserved CR3022 site. This antibody can block ACE2 without directly binding to the RBS. Such antibodies offer extraordinary potency against wide-ranging SARS-CoV-2 VOCs and sarbecoviruses.
Conclusions
In the current study, the authors have identified the vulnerable sites of the SARS-CoV-2 RBD, which can be targeted by antibodies. This could mitigate challenges associated with mutations that reduce antibody efficiency.
Journal reference:
- Yuan, M., Zhu, X., He, W., et al. (2022) A broad and potent neutralization epitope in SARS-related coronaviruses. Biophysics and Computational Biology. Biophysics and Computational Biology 119 (29) e2205784119. doi:10.1073/pnas.2205784119