
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
SARS-CoV-2, which is the pathogen responsible for the coronavirus disease 2019 (COVID-19) pandemic, is a single-stranded, positive-sense ribonucleic acid (RNA) virus belonging to the human betacoronavirus family.
SARS-CoV-2 has a 30-kilobase (kb) genome that encodes both structural proteins and NSPs. While structural proteins have important roles in viral infectivity, transmissibility, and virulence, NSPs are crucial for maintaining the viral life cycle inside host cells.
Among various NSPs, the most virulent is NSP1, as it causes global inhibition of host translational machinery, thereby leading to suppression of gene expression. Mechanistically, NSP1 inserts its C-terminal domain into the host ribosomal messenger RNA (mRNA) channel to inhibit translation. NSP1-mediated suppression of host gene expression eventually leads to inhibition of innate immune responses.
In the current study, scientists analyze the mutational changes in NSP1 sequences and determine the dynamics of its mutational landscape in various geographical regions.
Emergence of mutations in NSP1 sequence
A total of 5,966,020 NSP1 sequences were analyzed in the current study. No mutation was identified in 93% of the tested sequences, thus indicating high conservation of this protein. Among the remaining sequences, 6.3% had one mutation, 0.2% had two mutations, 0.003% had three mutations, and 0.001% had more than four mutations.
Considering various geographical regions, NSP1 sequences obtained from Europe had the highest frequency of one and two mutations as compared to those obtained from North America, South America, Asia, Oceania, and Africa.
In contrast, Oceania samples had the lowest frequency of one and two mutations in NSP1. A high frequency of three and more mutations in the NSP1 sequence was observed in samples obtained from Africa.
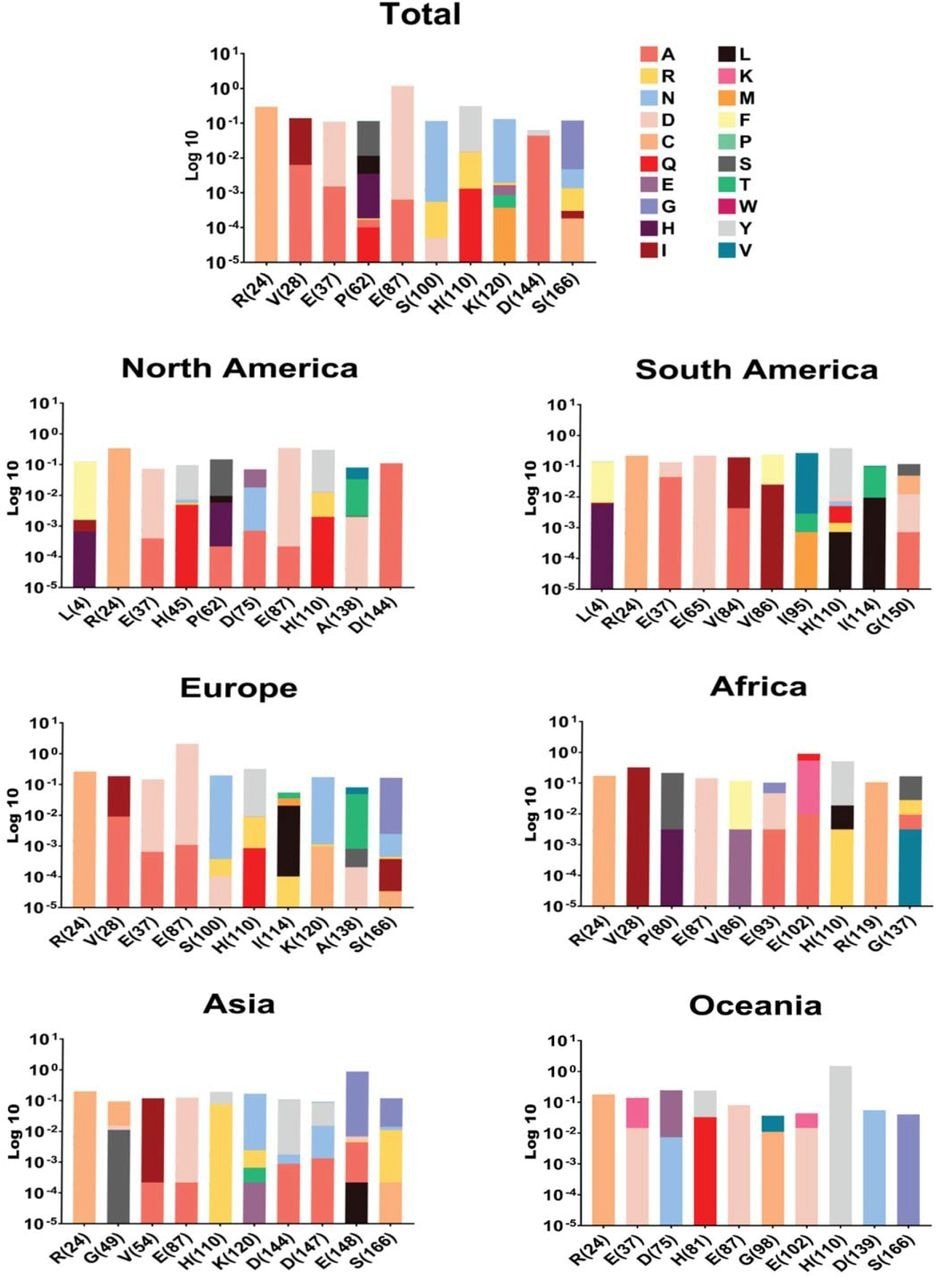
NSP1 top ten mutations with the highest frequency globally and geographic areas, including North America, South America, Europe, Asia, Oceania, and Africa. The position of changed amino acids and substituted ones is shown differently based on the logarithm 10 of data frequency in the percentage of substituted AAs. The mutation frequency was evaluated for each of them by normalizing the number of genomes carrying a given mutation in an intended geographic area.
High-frequency mutations in NSP1
The heat map of the NSP1 mutational landscape identified a specific amino acid segment of residues 72 to 126 that contains a high frequency of mutations. Thus, this segment was considered a mutational hot spot.
Among NSP1 mutations, E87D, H110Y, and R24C were identified as high-frequency mutations. More specifically, the frequency of E87D mutation was highest in Europe and lowest in Oceania.
The highest and lowest frequencies of H110Y mutation were observed in Oceania and Asia, respectively. Regarding the R24C mutation, North America had the highest frequency, whereas Africa had the lowest frequency.
These findings suggest that the frequency of NSP1 mutations varies between continents. Furthermore, these mutations may have a significant impact on COVID-19 severity, mortality, and drug or vaccine efficacy.
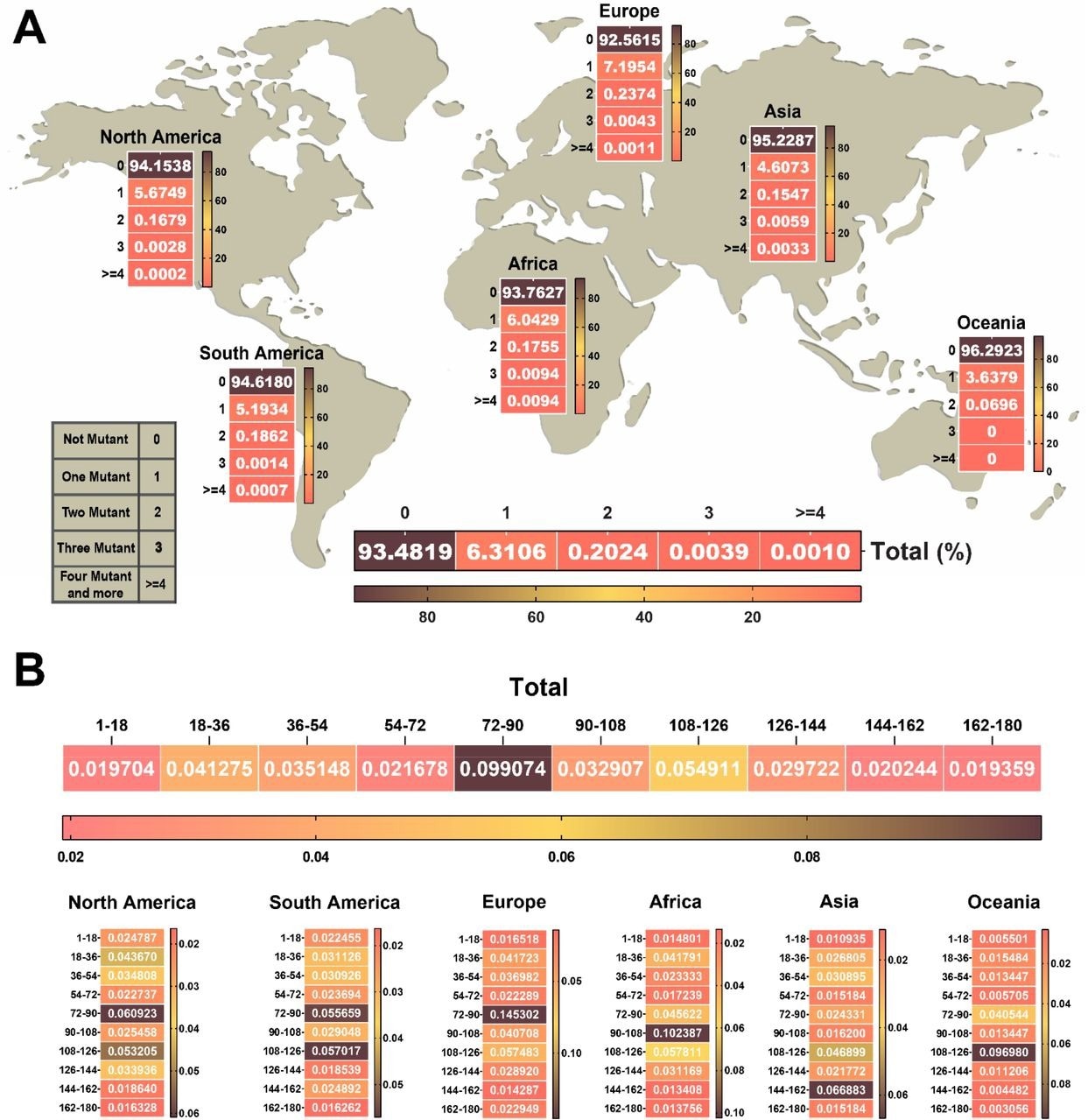 The heat map of the number and approximate region of mutations in NSP1 of SARS-CoV-2. (A) Heat map chart of the mutations’ number in NSP1 of SARS-CoV-2 until January 2022 in each continent. (B) The NSP1 protein was divided into ten segments. The rate of each mutation per segmented 18 amino acids was indicated in the heat map of the protein. The highest frequency rate occurred in the 72 to 126 amino acid sequence of the NSP1 protein that we can consider as a hot spot region in NSP1. The percentage of data were used to draw heat maps.
The heat map of the number and approximate region of mutations in NSP1 of SARS-CoV-2. (A) Heat map chart of the mutations’ number in NSP1 of SARS-CoV-2 until January 2022 in each continent. (B) The NSP1 protein was divided into ten segments. The rate of each mutation per segmented 18 amino acids was indicated in the heat map of the protein. The highest frequency rate occurred in the 72 to 126 amino acid sequence of the NSP1 protein that we can consider as a hot spot region in NSP1. The percentage of data were used to draw heat maps.
Dynamics of NSP1 mutation evolution
The analysis of mutational frequency in different geographical regions over a period of two years during the pandemic revealed that the frequencies of R24C and E87D mutations peaked in May 2020 and May 2021, respectively.
The frequency of the E87D mutation showed the highest level of fluctuation. In Europe, this mutation reached its peak in July 2021 and further increased in January 2022.
The analysis of the structural and functional impact of the E87D mutation revealed that the conversion of glutamic acid to aspartic acid at position 87 caused destabilization of the NSP1 structure. The mutation-induced alteration in intramolecular binding affinity could be responsible for structural destabilization.
Furthermore, the E87D mutation was found to increase the molecular flexibility of the NSP1 structure.
Implications
The current study describes the mutational landscape of the SARS-CoV-2 NSP1 sequence. Despite being highly conserved across coronaviruses, NSP1 genetic sequence may acquire mutations during viral evolution. Three notable mutations, including E87D, H110Y, and R24C have been identified in NSP1, with E87D increasing in frequency over time.
Given the impact of E87D on the NSP1 structure, further studies are needed to understand its functional impact on disease severity and therapeutic efficacy.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ghaleh, S. S. & Rahimian, K. (2022). SARS-CoV-2 Non-Structural Protein 1(NSP1) Mutation Virulence and Natural Selection: Evolutionary Trends in the Six Continents. bioRxiv. doi:10.1101/2022.07.22.501212. https://www.biorxiv.org/content/10.1101/2022.07.22.501212v1
- Peer reviewed and published scientific report.
Ghaleh, Samira Salami, Karim Rahimian, Mohammadamin Mahmanzar, Bahar Mahdavi, Samaneh Tokhanbigli, Mahsa Mollapour Sisakht, Amin Farhadi, Mahsa Mousakhan Bakhtiari, Donna Lee Kuehu, and Youping Deng. 2023. “SARS‐CoV‐2 Non-Structural Protein 1(NSP1) Mutation Virulence and Natural Selection: Evolutionary Trends in the Six Continents.” Virus Research 323 (January): 199016. https://doi.org/10.1016/j.virusres.2022.199016. https://www.sciencedirect.com/science/article/pii/S0168170222003458.