
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Most coronavirus disease 2019 (COVID-19) vaccines require prefusion-stabilized SARS-CoV-2 S protein as an immunogen for enhanced efficacy against this virus. K986P and V987P S mutations, for example, stabilize its pre-fused conformation.
All methods that are currently used to engineer prefusion-stabilized S-based vaccine immunogens involve tedious experimental work and rely on structural information. The structural determination and subsequent expression, purification, and characterization of each S mutation are laborious, thus hindering their quick identification.
Previous studies have shown that several other mutations could also improve SARS-CoV-2 S expression. For example, an S construct known as HexaPro containing F817P, A892P, A899P, and A942P mutations, in addition to K986P and V987P, has been shown to prove S expression.
About the study
In the present study, researchers describe a novel high-throughput approach to measure the fusogenicity of an array of SARS-CoV-2 S mutations in parallel, all of which could potentially stabilize its pre-fusion conformation without relying on structural information.
This method combines a fluorescence-based fusion assay, mammalian cell display technology, and deep mutational scanning (DMS). Moreover, it targets a conformational rearrangement in the S2 domain that covers the first heptad repeat (HR1), which comprises 912-984, and central helix (CH) of residues 985-1034.
The researchers constructed a DMS library comprised of amino acid mutations from residues 883 to 1034 of membrane-bound S protein and used it for encoding and expressing one S mutant in a HEK293T landing pad cell. All S-expressing cells also expressed mNeonGreen211 (mNG211) and were termed S-expressing cells.
A stable cell line that expresses human angiotensin-converting enzyme 2 (hACE2) and mNG21-10, which was named hACE2-expressing cells, was also developed.
The expression of membrane-bound S in 122 HEK293T landing pad cells and the formation of green-fluorescent syncytia due to the fusion of both types of cells was confirmed through flow cytometry (FC). The fusion assay was optimized to maximize the formation of green-fluorescent syncytia while minimizing the risk of clogging the cell sorter.
The fusion and expression scores for each of the 2,736 missense mutations, 152 nonsense mutations, and 152 silent mutations in the S protein were obtained. While a higher expression score indicated higher S expression, a higher fusion score indicated greater fusogenicity of the S protein. The expression and fusion scores were normalized so that the average score of silent mutations and nonsense mutations was one and zero, respectively.
Three and two biological replicates were used for the expression and fusion assays, respectively. An adjusted fusion score that represented the residual of a linear regression model of fusion score on expression score was also obtained.
Lastly, the researchers combined the validated fusion-incompetent mutations K986P, V987P, D994Q 181, and Q1005R to generate double, triple, and quadruple mutants of the membrane-bound or full-length S protein.
Study findings
Except for F817P, all other mutations in HexaPro present in the DMS library were screened. The DMS data consistently showed that A899P had minimal influence on S expression, whereas A892P and A942P markedly increased its expression.
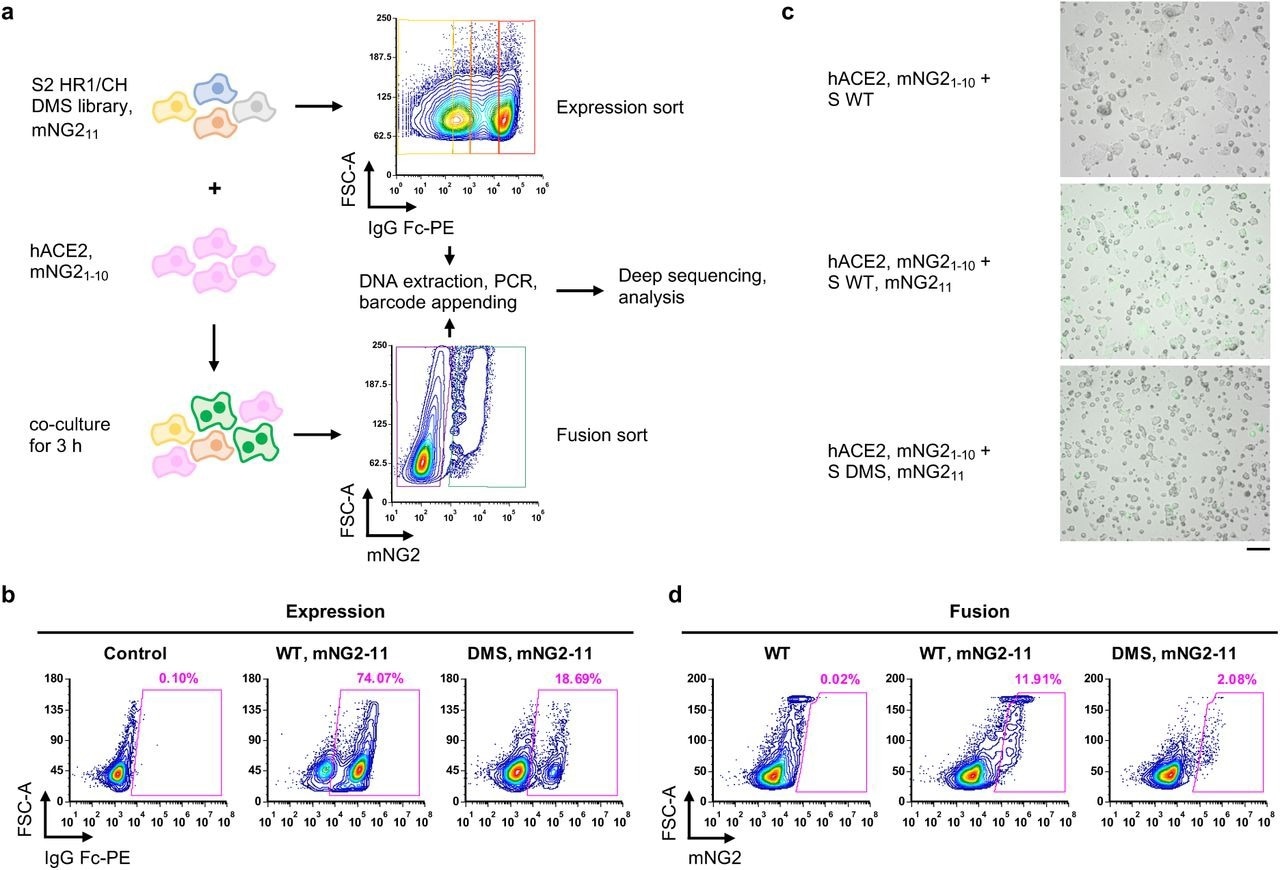 Measuring protein expression and fusogenicity of SARS-CoV-2 S mutations using deep mutational scanning. a, Schematic of high-throughput expression and fusion assays for S mutants. b, Flow cytometry analysis of S protein expression in HEK293T landing pad cells that encoded WT S or the DMS library. c, Fluorescent micrographs of co-culturing S-expression cells with hACE2-expressing cells. Scale bar: 100 μm. d, Flow cytometry analysis of fusion activity of co-culturing hACE2-expressing cells with HEK293T landing pad cells that encoded WT S or the DMS library. Components of split mNG2 are indicated where present.
Measuring protein expression and fusogenicity of SARS-CoV-2 S mutations using deep mutational scanning. a, Schematic of high-throughput expression and fusion assays for S mutants. b, Flow cytometry analysis of S protein expression in HEK293T landing pad cells that encoded WT S or the DMS library. c, Fluorescent micrographs of co-culturing S-expression cells with hACE2-expressing cells. Scale bar: 100 μm. d, Flow cytometry analysis of fusion activity of co-culturing hACE2-expressing cells with HEK293T landing pad cells that encoded WT S or the DMS library. Components of split mNG2 are indicated where present.
Among biological replicates, the Pearson correlation coefficient of expression scores ranged from 0.72 to 0.79, whereas that of fusion scores was 0.61, thus confirming the reproducibility of the DMS experiments.
The expression and fusion scores distribution of silent mutations were significantly different from those of nonsense mutations, thereby indicating that DMS experiments could distinguish mutants with different expression and fusogenicity levels.
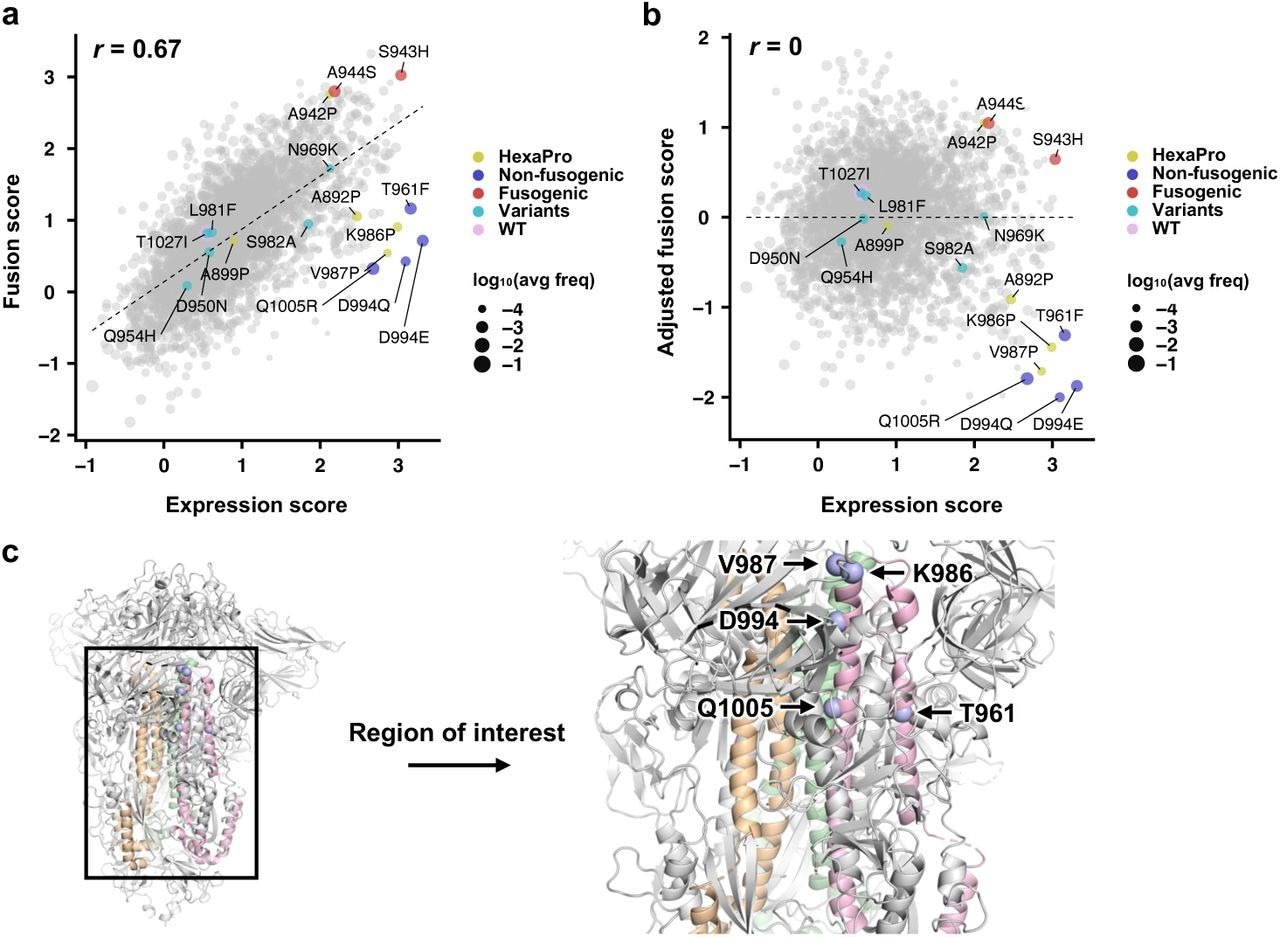 Expression and fusion scores of individual mutations in the DMS library. a, Plot of fusion score against expression score for each mutant is shown. WT is indicated in pink. Mutations used in HexaPro24 are in yellow. Representative fusion-incompetent mutations identified in this study are in purple (non-fusogenic). Representative mutations that enhance S fusogenicity are in red (fusogenic). Mutations found in major SARS-CoV-2 variants (Extended Data Table 1) are in teal (variants). Each data point represents one mutation in the DMS library. Individual data points are sized according to average frequency of the corresponding mutations. b, Plot of adjusted fusion score against expression score for each mutant is shown. Pearson correlation coefficient, r, is shown in a,b. c, Locations of fusion-incompetent mutations are indicated by light blue spheres. Regions that are mutated in the DMS library are colored wheat, green and pink for each monomer.
Expression and fusion scores of individual mutations in the DMS library. a, Plot of fusion score against expression score for each mutant is shown. WT is indicated in pink. Mutations used in HexaPro24 are in yellow. Representative fusion-incompetent mutations identified in this study are in purple (non-fusogenic). Representative mutations that enhance S fusogenicity are in red (fusogenic). Mutations found in major SARS-CoV-2 variants (Extended Data Table 1) are in teal (variants). Each data point represents one mutation in the DMS library. Individual data points are sized according to average frequency of the corresponding mutations. b, Plot of adjusted fusion score against expression score for each mutant is shown. Pearson correlation coefficient, r, is shown in a,b. c, Locations of fusion-incompetent mutations are indicated by light blue spheres. Regions that are mutated in the DMS library are colored wheat, green and pink for each monomer.
The fusion assay measured the fusogenicity at the cellular level, rather than at the single-molecule level. Moreover, the fusion score positively and consistently correlated with the expression score.
Mutations with a low adjusted fusion score and high expression score included K986P and V987P, both of which are used in current COVID-19 vaccines, thus demonstrating that the study method could identify prefusion-stabilizing mutations.
In addition to K986P and V987P, the study method identified other mutations in HR1 and CH which had low adjusted fusion and high expression scores, of which included T961F, D994E, D994Q, and Q1005R. Notably, D994E and D994Q clustered at the same amino acid residue positions and were chemically similar.
Consistent with the DMS data, the effects of T961F, D994E, D994Q, and Q1005R on S expression and fusogenicity were comparable to K986P and V987P in the validation experiments.
 Validation of candidate prefusion-stabilizing mutations. a-c, Expression of prefusion-stabilizing mutations (a), fusion-enhancing mutations (b), and combinations of candidate prefusion-stabilizing mutations of S (c) relative to WT. Of note, the numerical values of fold change in median fluorescence intensity (MFI) indicate relative and not absolute fold changes in surface expression levels of S. d-f, Fold change in fusion activity of candidate prefusion-stabilizing mutations (d), fusion-enhancing mutations (e), and combinations of candidate prefusion-stabilizing mutations of S (f) relative to WT at 3 hours post-mixing with hACE2-expressing cells. Abbreviations for combinatorial mutations are as follows: 2P, K986P/V987P; 2PQ, K986P/V987P/D994Q; 2PR, K986P/V987P/Q1005R; 2PQR, K986P/V987P/D994Q/Q1005R. Fold changes are shown as mean ± range. Data are from n = 2 independent replicates. Source data are available.
Validation of candidate prefusion-stabilizing mutations. a-c, Expression of prefusion-stabilizing mutations (a), fusion-enhancing mutations (b), and combinations of candidate prefusion-stabilizing mutations of S (c) relative to WT. Of note, the numerical values of fold change in median fluorescence intensity (MFI) indicate relative and not absolute fold changes in surface expression levels of S. d-f, Fold change in fusion activity of candidate prefusion-stabilizing mutations (d), fusion-enhancing mutations (e), and combinations of candidate prefusion-stabilizing mutations of S (f) relative to WT at 3 hours post-mixing with hACE2-expressing cells. Abbreviations for combinatorial mutations are as follows: 2P, K986P/V987P; 2PQ, K986P/V987P/D994Q; 2PR, K986P/V987P/Q1005R; 2PQR, K986P/V987P/D994Q/Q1005R. Fold changes are shown as mean ± range. Data are from n = 2 independent replicates. Source data are available.
Multiple HR1 and CH mutations, including those found in SARS-CoV-2 variants, did not negatively impact the expression or fusogenicity of the S protein. HR1 and CH were associated with high degrees of evolutionary conservation among betacoronaviruses.
Most SARS-CoV-2 neutralizing antibodies target the receptor-binding domain (RBD) region of the S protein; therefore, HR1 and CH may be under low selection pressure. Another explanation for this observation is that other evolutionary constraints on HR1 and CH might be present in vivo.
Although higher than the wild-type S protein, surface expression of S mutation combinations was comparable. None of these S mutation combinations fused with hACE2-expressing cells.
Size exclusion chromatography (SEC) of all mutants revealed that the Q1005R mutation likely increased the formation of higher order oligomers of soluble S ectodomain. Thus, specific S mutations could improve S expression in membrane-bound form but not soluble ectodomain form.
Conclusions
Given that prefusion-stabilization of S protein is critical for viral immunogen design, the study results have profound implications for COVID-19 vaccine development. Furthermore, the findings highlight that the evolutionary constraints of S2 are relevant to SARS-CoV-2 antigenic drift and the designing of universal coronavirus vaccines.
Nevertheless, future studies should continue to investigate the relationship between S protein expression, fusogenicity, and SARS-CoV-2 replication fitness to determine the biophysical processes that contribute to SARS-CoV-2 evolution.
The prefusion-stabilizing mutations of betacoronavirus S proteins were first reported in 2010, long before the COVID-19 pandemic began. This helped accelerate the development of COVID-19 vaccines, in addition to advancements in messenger ribonucleic acid vaccine technology.
The next pandemic could be due to a virus for which there is a lack of this type of information. Thus, maximizing the speed of vaccine immunogen design to accelerate vaccine development technology for yet-to-emerge viruses with the potential to cause pandemics is imperative.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tan, T. J. C., Mou, Z., Lei, R., et al. (2022). High-throughput identification of prefusion-stabilizing mutations in SARS-CoV-2 spike. bioRxiv. doi:10.1101/2022.09.24.509341. https://www.biorxiv.org/content/10.1101/2022.09.24.509341v1.
- Peer reviewed and published scientific report.
Tan, Timothy J. C., Zongjun Mou, Ruipeng Lei, Wenhao O. Ouyang, Meng Yuan, Ge Song, Raiees Andrabi, et al. 2023. “High-Throughput Identification of Prefusion-Stabilizing Mutations in SARS-CoV-2 Spike.” Nature Communications 14 (1): 2003. https://doi.org/10.1038/s41467-023-37786-1. https://www.nature.com/articles/s41467-023-37786-1.