Comparatively, T-cell immunity, which primarily targets the viral spike (S) protein, is less like to be affected by SARS-CoV-2 variants and, as a result, can remain effective in preventing severe symptoms associated with the coronavirus disease 2019 (COVID-19).
Study: Progressive loss of conserved spike protein neutralizing antibody sites in omicron sublineages is balanced by preserved T-cell recognition epitopes. Image Credit: Billion Photos / Shutterstock.com

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
Although most viral mutations do not significantly impact their properties, specific mutations can alter viral transmissibility, disease severity, and the effectiveness of therapeutic medicines, diagnostic tools, public health measures, and vaccines. Therefore, the threat of these types of mutations has led scientists worldwide to screen emerging and circulating SARS-CoV-2 variants continuously.
The SARS-CoV-2 Omicron variant has evolved into multiple sublineages, the earliest of which included BA.1, BA.2, BA.4, and BA.5. More recently, additional Omicron sublineages including BA.4.6, BF.7, BA.2.75, BA.2.75.2, BQ.1.1, and XBB have been identified and are currently emerging as dominant circulating strains to replace the previously dominant BA.5.
In addition to understanding these new variants' transmissibility, pathogenicity, and immune evasion properties, researchers have also examined the immunity patterns of human populations. These patterns are often determined by repeated infections with SARS-CoV-2 variants of concern (VOCs) and vaccination doses.
About the study
The researchers of the current study utilized their established artificial intelligence/machine learning-based early warning system to evaluate the immune responses of both naive vaccinated individuals, as well as those who have experienced breakthrough infections, against the Omicron sublineages BA.4.6, BF.7, BA.2.75, BA.2.75.2, BQ.1.1, and XBB.
Three study cohorts were assessed, including 18 infection-naïve individuals who received three doses of the Pfizer-BioNTech BNT162b2 messenger ribonucleic acid (mRNA) vaccine and were 55 years old age or younger. Fifteen individuals aged 60 years or older who received four BNT162b2 vaccine doses were also included.
Furthermore, triple-vaccinated individuals who experienced breakthrough infections with Omicron sublineages were included in the study. Fourteen of these individuals were infected with BA.1, 19 were infected with BA.2, and 17 were infected with BA.4/BA.5.
Serum neutralization was assessed by 50% pseudovirus neutralization (pVN50) geometric mean titers (GMTs). In addition, a total of 506 neutralizing B-cell epitopes within the spike protein were examined to determine the degree to which B-cell epitope conservation corresponds to individual SARS-CoV-2 variant cross-neutralization.
BA.2.75.2 and XBB escape neutralization by vaccination and Omicron infection
When the sera of individuals who received three and four vaccine doses were tested against BA.4/BA.5, GMT values were five- to six-fold lower than the wild-type strain. Similar GMT values were reported when the sera of vaccinated individuals with BA.1 infection were tested against BA.4/BA.5; however, a history of infection with BA.2 and BA.4/BA.5 led to higher GMT titers against BA.4/BA.5.
The neutralizing titers of all three patient groups who experienced breakthrough infection were more significant against Omicron BA.4.6/BF.7 and BA.2.75 compared to naive SARS-CoV-2 vaccine recipients. In addition, low titers were observed against BQ.1.1 when the sera of naïve and BA.1 convalescents were tested, whereas BA.2 and BA.4/BA.5 convalescents exhibited moderately higher neutralizing titers against this subvariant.
A history of breakthrough infection with BA.2 in vaccinated individuals led to greater cross-neutralization against BA.4/BA.5 and BA.4.6/BF.7 compared to triple-vaccinated individuals. This neutralization was even more significant after breakthrough infection with BA.4/BA.5, even when quadruple-vaccine recipients were considered.
These findings indicate that previous infection with some Omicron sublineages continues to protect against some of these subvariants. However, BA.2.75.2 and XBB have evolved to successfully evade neutralizing antibody responses in both naïve vaccinated individuals and those who have experienced breakthrough infection with previous Omicron subvariants.
B-cell and T-cell epitope conservation
Of the 506 neutralizing B-cell epitopes analyzed, 91% comprised a position altered by at least one SARS-CoV-2 variant. Although up to 43% of B-cell epitopes were partially conserved in the Alpha, Beta, and Delta variants, this conservation significantly declined to less than 22% in the Omicron BA.1 variant and less than 12% in BA.2.75.2 and XBB subvariants.
A total of 260 unique human leukocyte antigen (HLA) class I epitope sequences were analyzed, 244 of which were found in the SARS-CoV-2 wild-type spike protein. Of these epitopes, about 27% comprised a position mutated in at least one analyzed SARS-CoV-2 variant. Conversely, of the 468 HLA class II epitopes analyzed, 49% were located within a mutated region.
Nevertheless, about 90% of CD8+ and CD4+ T-cell epitopes were conserved in the Alpha, Beta, and Delta variants. Similarly, high rates of 80% and about 70% CD8+ and CD4+ T-cell epitope conservation were observed in the Omicron BA.2.75.2, BQ.1.1, and XBB strains.
This finding is consistent with previous evidence that describes the sustenance of T-cell immunity against Omicron subvariants in individuals with a history of SARS-CoV-2 wild-type infection.
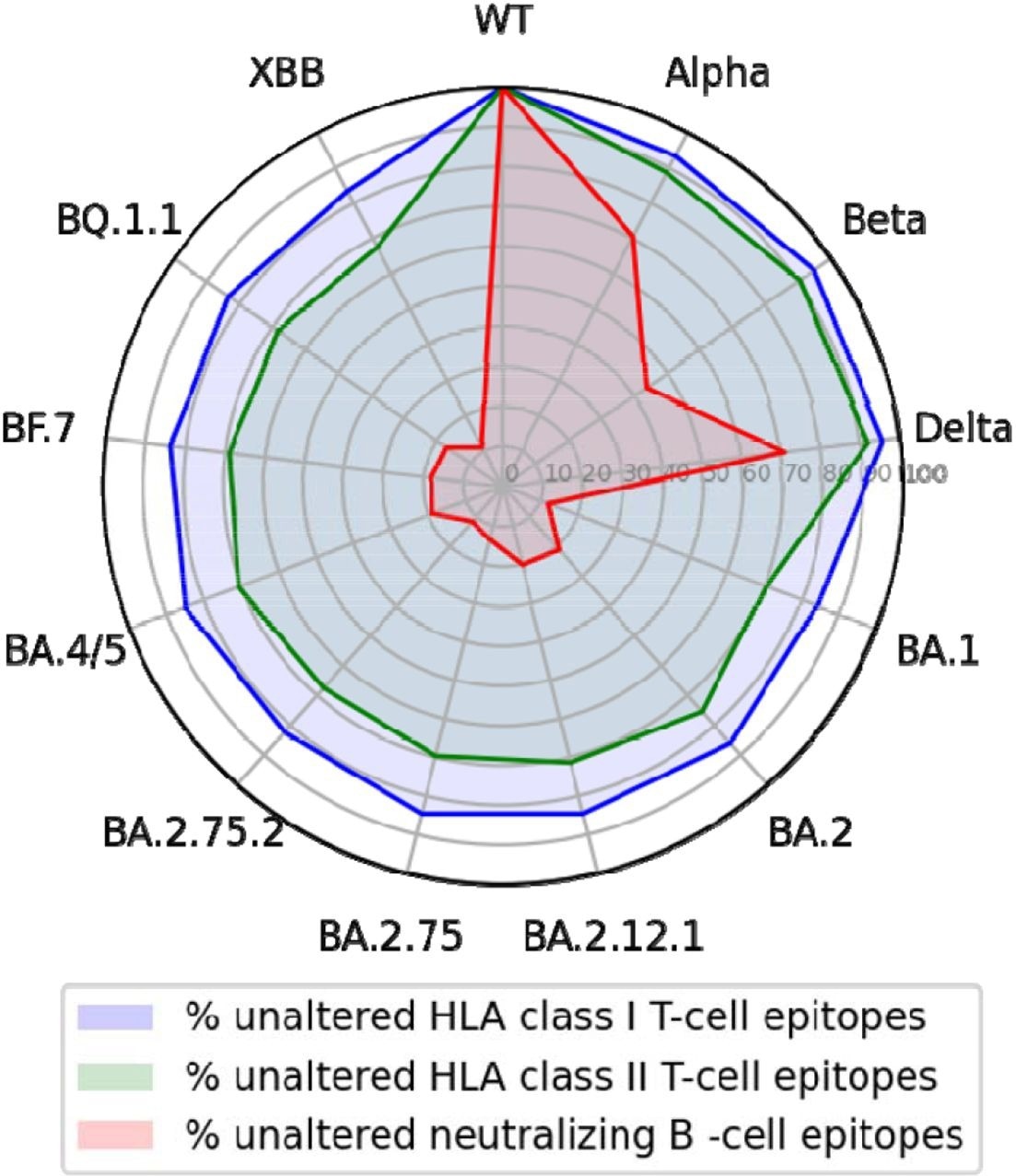
T-cell epitopes of the wild-type SARS-CoV-2 S glycoprotein but not epitopes for neutralizing antibodies are largely conserved across variants.
Conclusions
The study findings indicate that SARS-CoV-2 VOCs are less likely to breach T-cell-mediated immunity at the population level. Thus, T-cell immunity can effectively limit severe COVID-19 symptoms, even in the absence of neutralizing antibodies.
Taken together, these findings emphasize the importance of boosting T-cell immunity through targeted vaccine strategies that induce CD8+ T-cell immunity in addition to antibody production.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Muik, A., Lui, B. G., Diao, H., et al. (2022). Progressive loss of conserved spike protein neutralizing antibody sites in omicron sublineages is balanced by preserved T-cell recognition epitopes. bioRxiv. doi:10.1101/2022.12.15.520569