The global impact of novel zoonotic pathogen spillovers has been highlighted through the ongoing coronavirus disease 2019 (COVID-19) pandemic caused by the rapid outbreak of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus. In the past, similar effects were recorded during the HIV/AIDs and Spanish Influenza pandemics.
Due to climate change, increase in global population, urbanization, and intensification of agriculture, changes in rodent species assemblages have occurred. Unfortunately, these factors have also enhanced the frequency of zoonotic spillover events, which elevate the risk of the emergence of novel zoonotic pathogens from rodents. Generally, endemic zoonoses disproportionately affect those who come from weaker socio-economic backgrounds, live in close contact with animals, and have limited access to healthcare.
Rodents and bats cause the most significant number of zoonotic spillover events. Notably, among 2,220 extant rodent species, 10.7% are regarded as reservoirs of zoonotic pathogens. Increased probabilities of being zoonotic reservoirs are associated with short gestation times and early maturation. Rodents with these traits can thrive in human-dominated spaces, making it essential to describe rodent species assemblages and host-pathogen associations.
In a recent PLoS Neglected Tropical Diseases journal study, scientists synthesized rodent trapping studies published between 1964–2022. The studies were conducted across West Africa, which has been identified as a high-risk region for rodent-borne disease spillover.
About the Study
The current study conducts four main exercises, the first being the analysis of geographic sampling biases concerning land use classification and human population density. Second, to understand differences in reported host geographic distributions, the results of step 1 were compared to curated host datasets (IUCN and GBIF).
Third, CLOVER, a consolidated dataset, was used to contrast the identified host-pathogen associations. The main aim of this step was to detect discrepancies in rodent host-pathogen associations and estimate the fraction of positive assays for pathogens of interest. The last step involved the investigation of the spatial extent of current host-pathogen sampling within the gathered data. The goal was to detect regions of sparse sampling of pathogens within their host ranges.
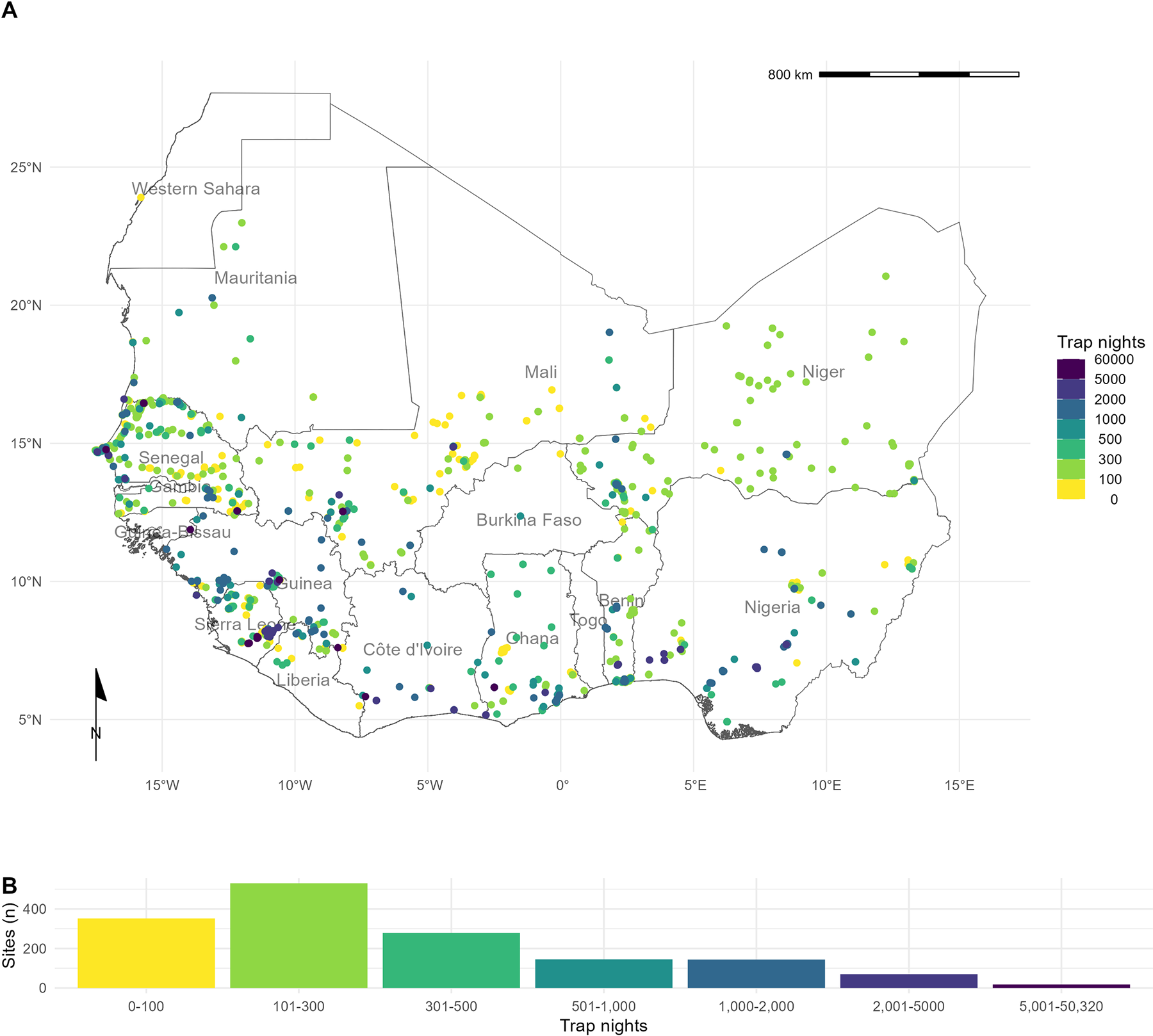 Rodent trapping sites across West Africa. A) The location of trapping sites in West Africa. No sites were recorded from Togo or The Gambia. Heterogeneity is observed in the coverage of each country by trap night (color) and location of sites. For example, Senegal, Mali, and Sierra Leone have generally good coverage compared to Guinea and Burkina Faso. B) Histogram of trap nights performed at each study site. A median of 248 trap nights (IQR 116–500) was performed at each site. A labeled map of the study region is attached in S5 Fig. Basemap shapefile obtained from GADM 4.0.4
Rodent trapping sites across West Africa. A) The location of trapping sites in West Africa. No sites were recorded from Togo or The Gambia. Heterogeneity is observed in the coverage of each country by trap night (color) and location of sites. For example, Senegal, Mali, and Sierra Leone have generally good coverage compared to Guinea and Burkina Faso. B) Histogram of trap nights performed at each study site. A median of 248 trap nights (IQR 116–500) was performed at each site. A labeled map of the study region is attached in S5 Fig. Basemap shapefile obtained from GADM 4.0.4
Key Findings
In the curated datasets, the majority of assayed rodents did not host known zoonotic pathogens. Twenty-five host-pathogen pairs were identified, of which 15 were not included in a consolidated host-pathogen dataset. The number and spatial extent of different species tested for a microorganism were limited, highlighting sampling bias.
Biodiversity data and, more generally, rodent trapping data showed significant spatial biases. The bias was greater in Guinea, Senegal, Benin, and Sierra Leone. Significant research has been conducted on the hazard of endemic zoonosis outbreaks (e.g., Lassa mammarenavirus) and the invasion of non-native rodent species (e.g., M. musculus and R. rattus).
Additional information on the human population density, trapping effort, and land use type have been incorporated, besides identifying prior rodent and pathogen sampling locations. This should help researchers identify locations where predictions based on the underlying data may be subject to sampling bias and also identify under-sampled locations. The modeling approach developed in this study identified North West Nigeria as an immediate priority for sampling rodents and their pathogens owing to the human-dominated landscape and the high population density. It was shown that a large part of West Africa is still under-sampled, particularly Burkina Faso, Côte d’Ivoire, Ghana, and Nigeria.
Rodent trapping studies provide data on both species detection and non-detection, which can improve models of species distributions. This is important to evaluate the effect of climate change and land use on the risk of zoonotic spillovers to human populations. However, currently available consolidated datasets, such as CLOVER, EID2, and GMPD2, do not comprise temporal or spatial components. Therefore, current models dependent on these data sources cannot account for spatial heterogeneity in pathogen prevalence.
Due to data sparsity, the temporal change over the six decades of rodent trapping studies could not be accounted for. It is possible that land use and population density at trapping sites varied over this time period. Despite this limitation, the finding that trapping is biased towards human-dominated and densely populated landscapes is expected to stand.
Conclusions
Under-studied regions should be prioritized in future rodent trapping studies. More information on rodents in West Africa should help in modeling the changing endemic zoonosis risk and the potential for the emergence of novel pathogens. More widespread sharing of research and data on locations of trapping sites and trapping efforts should be highly encouraged. The researchers hypothesized that curated datasets, supplemented with rodent trapping studies, could be vital in quantifying the risk of zoonotic spillover events and tracking the emergence of new pathogens.