
 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
About 10% of patients who have recovered from COVID-19 suffer post-acute sequelae of SARS-CoV-2 infection (PASC), often referred to as LC. Although researchers have proposed multiple mechanisms that may contribute to LC, there remains an urgent need to understand the pathogenesis of this condition better, as LC cases continue to surge worldwide and increase the burden on public healthcare systems.
Recently, omics-based approaches performed on plasma samples of LC patients have shown elevated levels of inflammatory cytokines, such as interferon-beta (IFN-β) and interleukin-8 (IL-8), as well as reduced cortisol levels.
Serological analyses have also identified elevated levels of SARS-CoV-2 autoantibodies in LC patients; however, these observations have not been consistently reported. In addition, single-cell transcriptomics of peripheral blood mononuclear cells (PBMCs) have identified increased levels of specific LC phenotypes, including myeloid and natural killer (NK) subsets.
T-cells, especially clusters of differentiation 4 (CD4)+ T-cells, which play a key role in adaptive immunity, are crucial in mounting an effective immune response to SARS-CoV-2. Nevertheless, studies assessing the impact of CD4+ cells during COVID-19 have provided conflicting results. To this end, while some studies have identified elevated SARS-CoV-2-specific T-cell responses in LC patients, others have observed a more rapid decay of SARS-CoV-2-specific CD8+ T-cells in this patient population.
About the study
In the present study, researchers used CyTOF for deep phenotypic characterization of T-cell immunity in LC patients. Next, these data were combined with standardized serological analyses and ribonucleic acid sequencing (RNAseq) and high-dimensional (HD) plasma proteomics-based proximity extension assay (PEA), both of which are omics techniques that enable the simultaneous quantitation of 384 plasma analytes.
This analysis was conducted over eight months and included both LC and a matched set of non-LC individuals. This allowed for the identification of unique LC-related immune features to provide important insights into the mechanistic aspects of this multifaceted debilitating disease.
For both LC and non-LC individuals, the researchers comprehensively monitored the expression IFNγ, IL-2, macrophage inflammatory protein 1b (MIP1b), tumor necrosis factor-alpha (TNF-α), and IL-6.
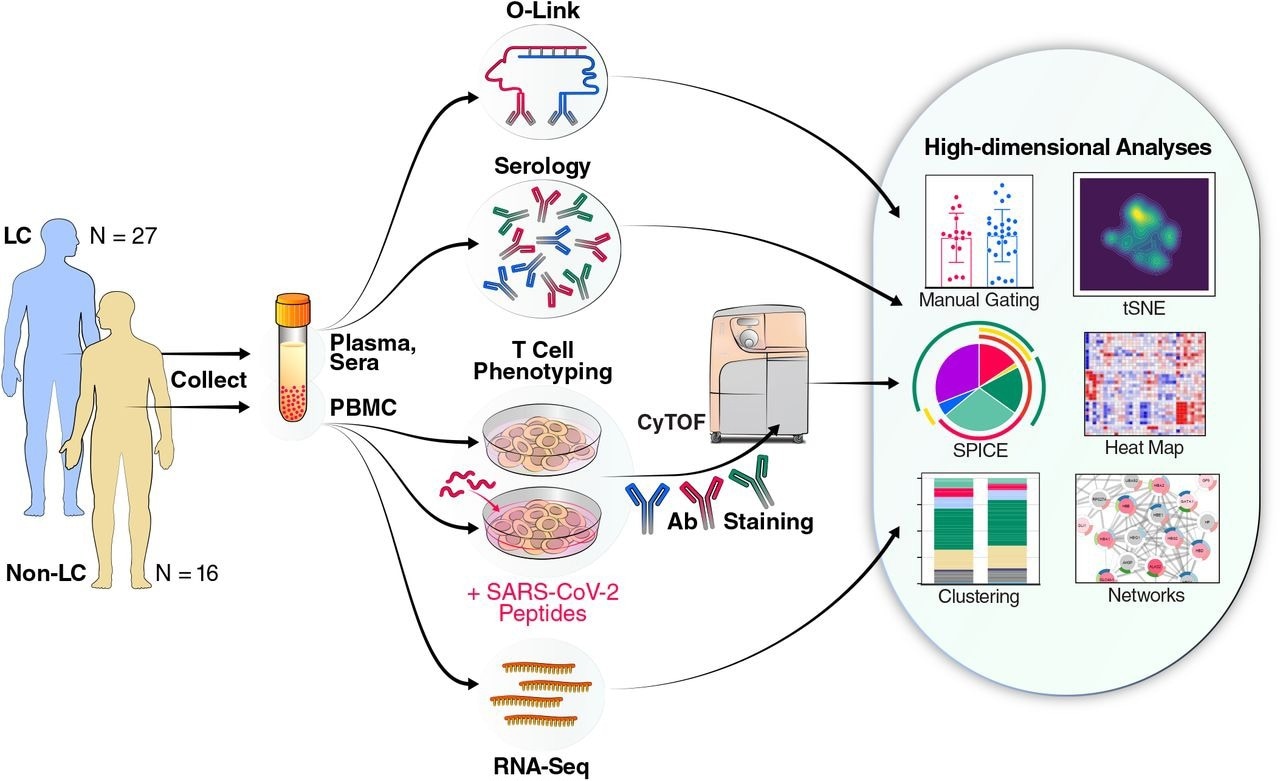
Schematic of experimental design and data analyses. Plasma and sera from 27 individuals with Long COVID (LC) and 16 individuals without LC (Non-LC) were subjected to Olink and serological analyses. PBMCs from the same individuals were subjected to RNAseq analysis, as well as to CyTOF analysis at baseline, or following a 6-hour stimulation with peptides derived from SARS-CoV-2 spike proteins (see Methods) to analyze T cell responses. The cells for CyTOF were treated with viability marker, fixed, and stained with a 39-parameter panel (Table S2) prior to analysis on a CyTOF instrument. The indicated tools on the right were then used for analyses of the resulting high-dimensional datasets.
Study findings
CyTOF data uncovered significant variations in total CD4+ T-cell counts in LC patients, with a significantly higher proportion of CD4+ central memory T-cells (TCM), T follicular helper cells (Tfh), and regulatory T-cells (Tregs). Notably, these cells may not necessarily be against SARS-CoV-2 but may be directed towards other viruses like herpes or certain auto-antigens.
Nevertheless, one recent study documented that people with increased levels of activated Treg cells during acute COVID-19 experienced LC about three months after recovering from the infection. This observation, along with the aforementioned finding of the current study, suggests that Tregs initiate and maintain LC.
LC patients in the current study did not harbor more polyfunctional anti-SARS-CoV-2 T-cells in the CD4 and CD8 compartments.
Anti-SARS-CoV-2 T-cells were detected in both LC and non-LC patients for several weeks after recovering from the infection. The persistence of TCM cells triggered by SARS-CoV-2 indicated the prolonged existence of a long-lived viral reservoir in body tissues.
LC-infected individuals had fewer anti-SARS-CoV-2 CD8+ T-cells but not overall CD8+ T cells that expressed the exhaustion markers programmed death 1 (PD1) and cytotoxic T-lymphocyte–associated antigen 4 (CTLA4) consistently, which is likely due to continuous stimulation with SARS-CoV-2 antigens.
Interestingly, the individuals with the highest T-cell exhaustion markers were not those with the most anti-SARS-CoV-2 antibodies, thereby suggesting that multiple LC endotypes might be responsible for persistent virus levels. Thus, future studies should evaluate other antivirals, apart from nirmatrelvir-ritonavir, that might reinvigorate the ability of T-cells to eradicate residual viremia.
One subset of cytotoxic and stimulated T-cells was amplified to a greater extent in females with LC, while males showed a contrasting pattern. A correlation between cytotoxic T-cells and gastrointestinal (GI) LC symptoms was also observed. Taken together, these findings indicate that future studies are needed to establish whether LC-related cytotoxic T-cell function varies with gender and GI symptoms.
Some LC patients produced IL-6 upon stimulation by the SARS-CoV-2 spike protein, which suggests that an inflammatory response against the virus persisted for at least eight months post-infection. Thus, future studies should investigate potential LC therapeutic strategies targeting IL-6.
LC patients also lost coordinated humoral and cellular immune responses to SARS-CoV-2 that exist in fully recovered individuals. Moreover, the PEA analysis revealed that the dysregulated production of IL-4 and IL-5 by Th2 cells likely thwarted humoral and cellular immune responses. Upstream initiators of these biomarkers could include persistent SARS-CoV-2 reserves, reactivation of viral co-infections, and autoimmunity.
About half of the individuals with LC with insignificant SARS-CoV-2 antibody levels exhibited significant SARS-CoV-2-directed T-cell responses. Inopportune crosstalk between B- and T-cells might contribute to the etiology of LC, which was supported by RNAseq data in this study.
Furthermore, LC patients exhibited co-upregulated expression of genes responsible for T-cell functionality and immunoglobulin synthesis. For example, LC patients had elevated SGALS9 gene expression, which resulted in more Galectin production and, as a result, greater cytokine release and COVID-19 severity.
LC patients with the most exhausted anti-SARS-CoV-2 CD8+ T-cell levels had the lowest frequencies of anti-SARS-CoV-2 CD4+ Treg cells, thus indicating their potentially faulty immune regulatory mechanisms. In addition, LC patients had upregulated gene expression of olfactory sensing and heme synthesis, which suggests the involvement of non-immune mechanisms in LC pathophysiology.
Several studies have reported that SARS-CoV-2 can bind hemoglobin to disturb heme metabolism in people with acute infection. Thus, future studies are needed to identify associations between iron metabolism and its relationship to coagulopathy in LC patients.
Conclusions
The study findings point to a dysregulated but highly pro-inflammatory signature in LC. While LC pathophysiology appears to be highly complex, the current study provided important insights into its potential pathophysiological contributing factors.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.