Herpesviruses are significant human and animal pathogens. Exploring the role of specific genes, understanding the pathogenesis of herpesviruses, and generating novel anti-viral vaccines and therapies all require the manipulation of the vast genome. The technique of BAC has greatly improved the power of herpesviruses studies to edit virus genomes. In recent years, progress has been made in BAC-based genome manipulation and screening procedures of recombinant BACs, which has facilitated the research of the herpes virus.
Herpesvirus genome editing methods based on BAC
RecA recombination method
RecA recombination is a bacterial endogenous homologous recombination system consisting of RecA recombinase and associated accessory proteins. RecB also possesses exonuclease activity, facilitating the formation of 30-end single-stranded deoxyribonucleic acid (ssDNA). RecA binds to ssDNA sites created by exonucleases and initiates the process of recognizing homologous areas on the BAC for future annealing and interaction. When a successful interaction is formed, nucleoprotein filaments capture double-stranded DNA (dsDNA) and exchange strands to generate heteroduplex DNA.
While the RecA-mediated homologous recombination approach for editing herpesvirus genomes present in eukaryotic cells is more effective than conventional methods, it has considerable limitations. Due to the existence of repetitive sequences in the genomes of the herpes virus, one key issue is that RecA expression can generate instability in the herpes virus's BAC clone, which can result in viral genome mutations.
λ-red recombination procedure
The λ-red recombination system comprises Exo, Beta, and Gam proteins and is generated from the phage. Gam protein may impede the Rec BCD binding to the dsDNA ends, inhibiting RecBCD exonuclease and restricting the breakdown of exogenous dsDNA. Exo proteins can interact with the terminals of dsDNA donors with homologous sequences flanking the target gene. Also, Beta protein is essential for the λ-red homologous recombination process. Being a protein that binds to single-stranded DNA, the beta protein interacts with the ssDNA generated by the Exo protein. Beta protein facilitates donor DNA fragment annealing and the homologous sequence observed at the target site of the replication of herpes virus BAC. Homologous recombination concludes with DNA replication.
Base editing method
Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas 9 is a powerful genome editing technology that has been applied effectively to a wide variety of eukaryotic organisms, including embryonic stem cells, human cell lines, mice, Drosophila, and Arabidopsis. Using CRISPR/Cas 9 gene editing technology with precise base editing technology, scientists have recently developed a method for effective gene editing involving a single guide ribonucleic acid (sgRNA )and a complex containing modified Cas9 proteins, a uracil glycosylase inhibitor (UGI), and cytosine deaminases.
In contrast to wild-type Cas9 proteins, the modified Cas9 lacks endonuclease activity and is catalytically inactive. Cytosine deaminase transforms a particular cytosine (C) site into uracil (U). At this stage, uracil glycosylase inhibitors can block the excision of intermediate product U, boosting the effectiveness of C to T conversion on the DNA chain and enabling single-base precise editing of G to A and C to T.
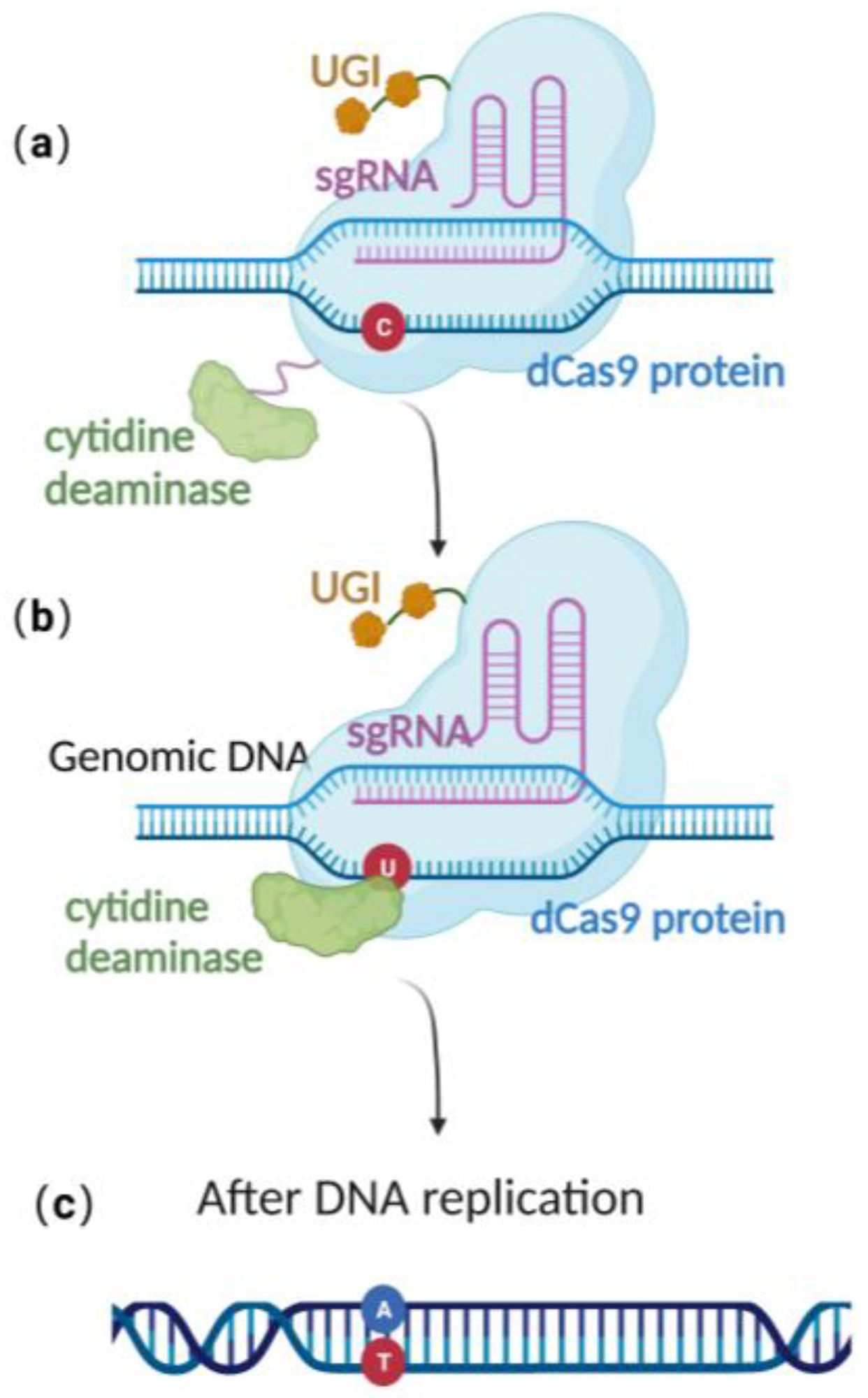
Schematic diagram of base editing method: (a) dCas9 mediates targeting without DSB formation; (b) cytosine deaminase converts C to U; (c) single-base precise editing is complete with the replication of DNA.
Techniques for screening herpesvirus mutants
Single-selection cassette
In the single selection cassette technique, along with homologous arms present on both sides of the target site as well as the sequence to undergo insertion, the donor typically adds selection cassettes, like resistance genes, in order to select designed BAC clones. Screening performed with the relevant marker gene phenotype significantly reduces the background of cells that have not undergone recombination, resulting in a screening effectiveness of greater than 95%. However, marker gene insertion into a target gene may impair the function of genes both downstream and upstream of the targeting location.
Combining a selection cassette with a site-specific recombinase recognition motif
For editing herpesvirus genomes, site-specific recombinase systems, including the yeast-derived flippase (Flp)-flippase recognition target (FRT) system and the P1 phage-derived Cre-locus of X-over P1 (loxP) system are often employed. During the editing process, the donor must incorporate codirectional LoxP/FRT regions on the two sides of the selection cassette. Recombinase enzyme expression eliminates the selection cassette positioned between the two sites. While this strategy effectively eliminates the lengthy selection cassette with a near-perfect success rate, it leaves a single LoxP or FRT site at the intergenic location of BAC insertion and excision.
Conclusion
The study findings showed that using BAC recombinant technology has remarkably broadened the future of herpesvirus study. However, the effectiveness of BAC-based gene editing still needs further development. The researchers believe that developing more efficient viral genome editing technologies based on BAC would enhance the precision and efficacy of genome editing while reducing the testing time and resources required.
Journal reference:
- Hao, Mengling, Jiabao Tang, Shengxiang Ge, Tingdong Li, and Ningshao Xia. 2023. “Bacterial-Artificial-Chromosome-Based Genome Editing Methods and the Applications in Herpesvirus Research.” Microorganisms 11 (3): 589. https://www.mdpi.com/2076-2607/11/3/589