Microbiomes have taken on major importance as producers and modulators of the host metabolism and homeostatic machinery. In a new study, researchers at the University of Calgary, Canada, explored the metabolomes of mice exposed to different types of microbes to understand the effects and the possible influences of age and sex on these outcomes.
The gut handles all ingested foods for further digestion and assimilation. In addition, it harbors a wealth of microbes that produce metabolic byproducts with multiple effects on host physiology. Therefore, changes in the gut microbiome's composition can shift the gut's metabolome as well.
For instance, the human gut microbiota produces a variety of vitamins and ferments undigested carbohydrates and proteins to produce short-chain fatty acids (SCFAs), lactate, and bile acids, among other molecules, affecting the immune and coagulation systems, as well as modulating the neuroendocrine network and influencing the gut-brain axis. These metabolic products may cause alterations in the type and abundance of metabolites absorbed via the epithelial barrier.
The use of germ-free mouse models (GFMM) is a recent advance in the study of microbial effects on the gut metabolome. It enables their separation from the effects of host metabolites. The current paper, published in the journal Nature Communications, made use of gnotobiotic mouse models, along with GFMM and specific pathogen-free mice (SPF).
The gnotobiotic mice (OMM12) were colonized with a selected set of 12 commensals from each of the five major phyla found in the mouse gut. These include Bacteroidota, Bacillota, Pseudomonadota, Actinomycetota and Verrucomicrobiota.
The researchers studied the metabolites in the peritoneal fluid, serum, liver, spleen, and urine, of mice at various ages, from just-weaned infants to both male and female adults.
What did the study show?
The findings show that the host metabolome is driven most notably by body size, modulated by microbiota, age, and sex, in that order.
The gut shows different regions with distinct metabolite signatures, with the most significant similarity being between the upper gastrointestinal (GI) and lower GI tract of GF vs colonized mice, respectively.
The upper GI tract (GIT) had higher concentrations of amino acids from the diet, such as tryptophan, leucine, and histidine, which decreased downstream, probably due to absorption. Sugars, fatty acids, and nucleosides were present more abundantly in the lower GIT due to microbial breakdown of proteins, carbs, and nucleic acids. This was reflected in the significantly higher concentrations of these digestion products in GF mice, which lack any microbes.
The most crucial factor in determining the differences in the metabolite concentrations at each site within the gut was the microbiota.
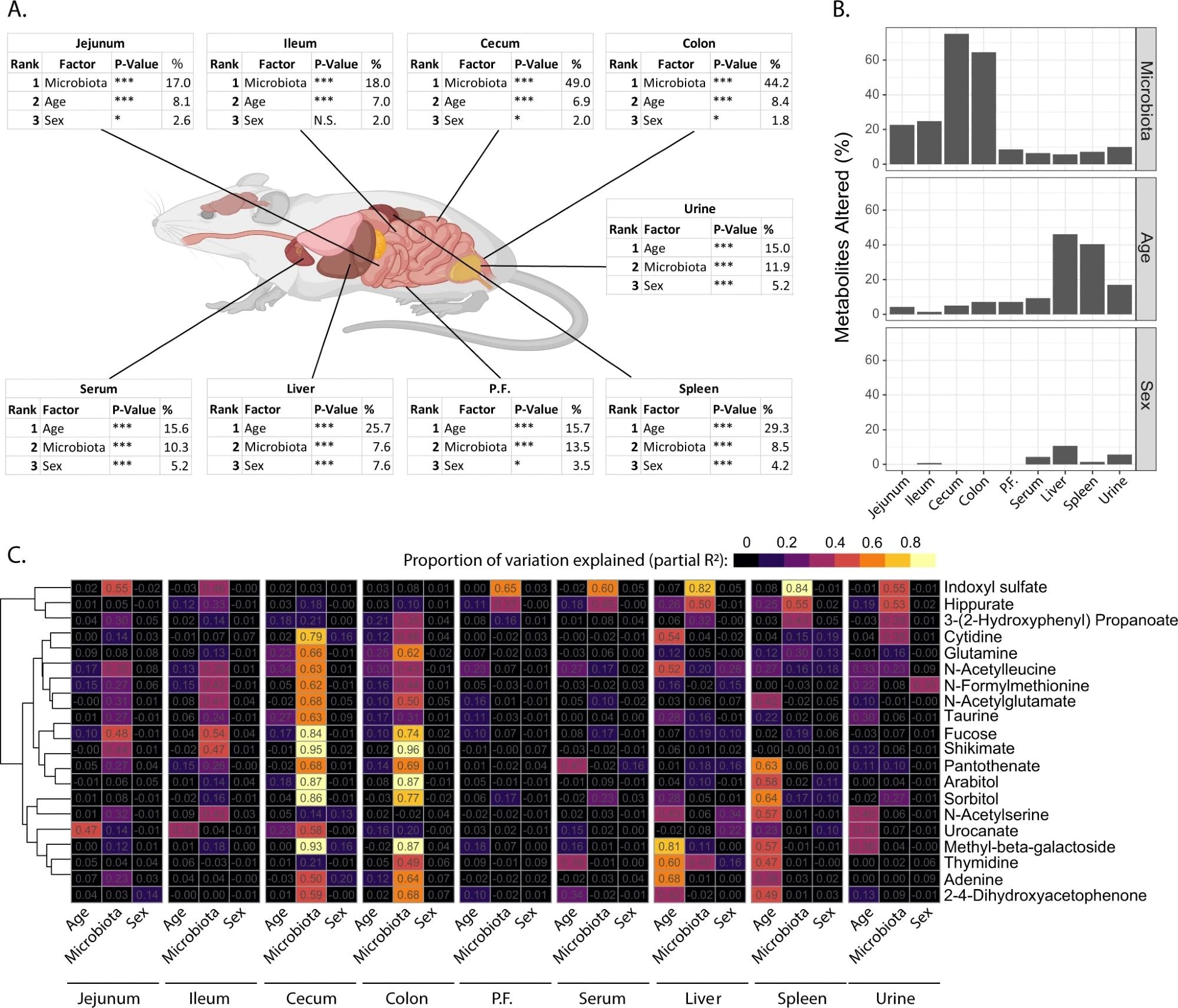 A Estimated proportion of variation explained by microbiota, age and sex in each site sampled based on PERMANOVA statistics. B Proportion of metabolites altered (PAdj <0.05) by microbiome, age, and sex at each site. C Partial R2 showing the relative contribution of age, microbiota, and sex in explaining the variation in metabolite abundance at each site. Metabolites shown are the 20 metabolites where the most variation was explained by the three factors, full heatmap for all metabolites is shown in Supplementary Figure 1. Data is representative of n = 72 samples / site (equal representation from male and female mice, GF, OMM12 and SPF colonized mice and 3-, 8- and 12-week-old mice). P.F. = peritoneal fluid. Parts of Fig. 2A were generated using Biorender.org under license. Source data are provided as a Source Data file.
A Estimated proportion of variation explained by microbiota, age and sex in each site sampled based on PERMANOVA statistics. B Proportion of metabolites altered (PAdj <0.05) by microbiome, age, and sex at each site. C Partial R2 showing the relative contribution of age, microbiota, and sex in explaining the variation in metabolite abundance at each site. Metabolites shown are the 20 metabolites where the most variation was explained by the three factors, full heatmap for all metabolites is shown in Supplementary Figure 1. Data is representative of n = 72 samples / site (equal representation from male and female mice, GF, OMM12 and SPF colonized mice and 3-, 8- and 12-week-old mice). P.F. = peritoneal fluid. Parts of Fig. 2A were generated using Biorender.org under license. Source data are provided as a Source Data file.
However, the effect of age was comparable to that of microbiota in the variation of metabolites in serum, urine, and peritoneal fluid. Age played a more prominent role in modulating the metabolite profile in the liver and spleen. Sex differences caused the most minor effect.
The metabolome in the three mouse models also showed the influence of microbial density and complexity. In the lower GIT, the greatest effect was due to the density of the microbiota, with the OMM12 showing changes in approximately half the metabolites. In contrast, the SPF mice showed changes in over 60% of metabolites in the colon and over half in the cecum.
Bacterial density is highest in these regions, accounting for the significant contributions of the microbiota to the host's metabolism. Still, the whole GIT showed a marked effect on microbial metabolism.
Different microbiota composition was reflected in varying metabolite profile. In the upper GIT, SPF and OMM12 mice had higher cholate levels compared to GF mice, but only SPF mice had higher taurocholate levels. Similarly, microbes in the SPF mice used up allantoin and raffinose in the lower gut, producing glutamine, nicotinate, and guanine, but OMM12 microbes used up only raffinose and produced only nicotinate.
Raffinose is composed of glucose, fructose, and galactose. The enzyme required for its breakdown, α-galactosidase, is found in microbes but not humans or mice. Nicotinate is a vital micronutrient required for the energy cycle of the cell and is derived from the amino acid tryptophan by microbes, apart from its availability as such in the diet.
"The abilities of the SPF and OMM12 microbiotas to similarly affect the abundance of molecules such as raffinose and nicotinate suggests that these may be well conserved metabolic pathways in bacteria, or that a specific member of the OMM12 consortia is able to normalize to the levels of SPF mice."
However, none of the 12 commensals in OMM12 mice appear to produce tryptophanase.
Similar changes were seen at other non-GI sites as well, indicating the "metabolic preferences of microbial communities, which has an impact on metabolite concentration."
The effects of age on microbial-induced metabolite profiles were also observable, though this was also modulated by the local microbiome. GF mice showed high levels of uridine and histidine, among others, in the lower GIT at eight weeks, compared to three weeks. This was not the case when comparing mice with different microbiota, namely, SPF and GF mice.
This indicates that the effect of the microbiota on host metabolism varies with the stage of development. Some metabolites also showed significant variation with sex in OMM12 or SPF mice but not GF mice, showing the metabolic differences affected by sex hormones in adult mice mediated by the microbiome.
The study helps to understand more about how microbes in the gut interact with the host via metabolites and could assist future research. For instance, it shows the significant effect of the specific GI site on the metabolome, defined chiefly by the microbiota.
The absence of multiple metabolic pathways in GF mice underlines the considerable contribution of the microbiome to human metabolism.
What are the implications?
"Leveraging microbe-metabolite-host interactions towards personalized medicine holds great promise for improving health and preventing disease."
The study shows that microbes differentially alter the levels of specific metabolites depending on the presence of certain taxa. In addition, age and sex modulated the effects of the microbiome on metabolism.
While the study was able to identify differences in 140 metabolites using semi-targeted methods, this is only the tip of the iceberg. Multiple orders of magnitude of metabolites remain that have not been identified by these methods. The study may thus provide a pattern for future microbiome studies concerning these modulators.