A new study uncovers how gut bacteria and blood metabolites signal early diabetes risk and how tailored diet and exercise could reverse the trend.
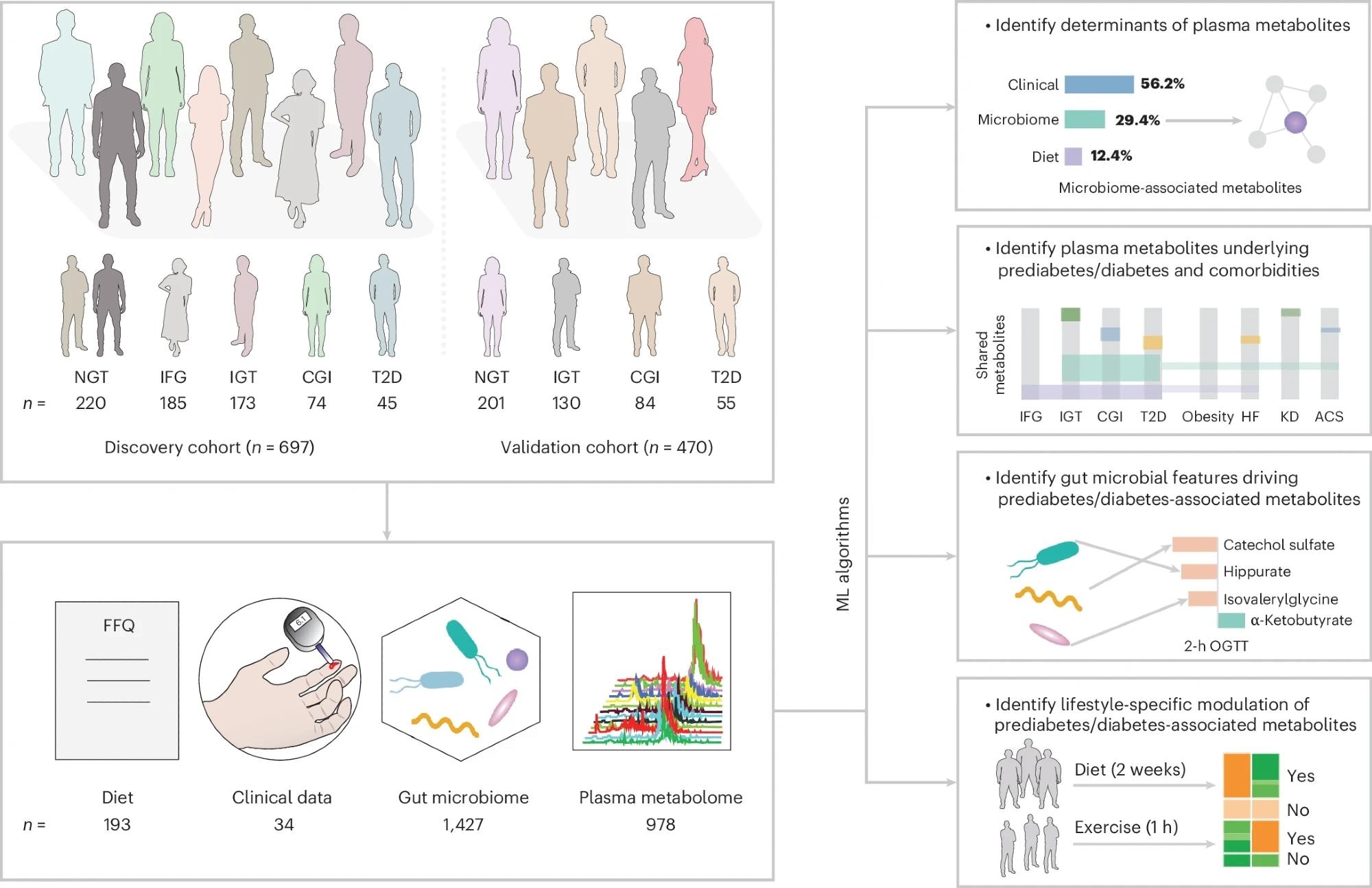 FBG and OGTT were used to screen individuals with varying degree of glucose intolerance. The GBDT algorithm was used to predict plasma metabolites based on collected data from the FFQ, clinical tests and gut microbiome profiling. n indicates the sample size for the two cohorts, or the number of features in the diet, clinical, gut microbiome and plasma metabolome datasets.
FBG and OGTT were used to screen individuals with varying degree of glucose intolerance. The GBDT algorithm was used to predict plasma metabolites based on collected data from the FFQ, clinical tests and gut microbiome profiling. n indicates the sample size for the two cohorts, or the number of features in the diet, clinical, gut microbiome and plasma metabolome datasets.
In a recent study in the journal Nature Medicine, researchers conducted a metabolome profiling study to investigate the role of microbial metabolites in prediabetes and type 2 diabetes (T2D). They used two Swedish cohorts comprising 1,167 participants aged 50–64 years for their analyses.
Study findings revealed the presence of 502 blood metabolites linked to impaired glucose homeostasis, 143 of which were associated with the human gut microbiome. The study highlights the role of microbiome-metabolome dynamics in prediabetes and T2D pathophysiology and the role of short-term lifestyle changes (diet and exercise) in modulating these dynamics.
Background
Type 2 diabetes (T2D) is a global public health concern, estimated to affect more than 830 million adults. The condition is chronic, characterized by the body’s inability to regulate glucose metabolism adequately, resulting in excessively high blood sugar levels, potentially leading to complications including cardiovascular diseases (CVDs), kidney diseases, nerve damage, and increased mortality risk.
Alarmingly, the prevalence of T2D is growing at unprecedented rates, rising from 200 million in 1990 to more than 830 million in 2022. Research has revealed that the condition’s pathophysiology is highly complicated, arising from the interplay between genetic and environmental variables. Recent studies have suggested the extensive role of diet and the gut microbiome in T2D pathogenesis, with an estimated 70% of T2D incidence now attributed to suboptimal diets and their adverse effects on gut bacteria.
Unfortunately, the mechanistic influence of gut microbial metabolites on T2D pathogenesis and progression remains poorly understood.
About the Study
The present study aims to address these gaps in the literature by identifying gut microbial metabolites modulating host (human) glucose control and, in turn, contributing to prediabetes and T2D. Study data were obtained from two Swedish prediabetes cohorts – the Impaired Glucose Tolerance (IGT; n = 697) cohort, which served as the ‘discovery’ cohort, and the Swedish CArdioPulmonary bioImage Study (SCAPIS; n = 470), which served as the ‘validation’ cohort.
Study data collection included morning fasting glucose measurements, a 75g oral glucose tolerance test (OGTT), and fasting venous blood sample collection. The results from these tests, in tandem with the 1999 World Health Organization criteria, were used to divide study participants into five subgroups: 1. Normal glucose tolerance (NGT), 2. Isolated impaired fasting glucose (IFG), 3. Impaired glucose tolerance (IGT), 4. Combined glucose intolerance (CGI), and 5. T2D.
Collected blood samples were subjected to plasma metabolomics using the Metabolon platform. Microbially associated metabolites were identified using gradient-boosting decision trees and random forest machine learning (ML) models.
Additionally, participants were required to complete FINDRISC questionnaires (which reflect insulin resistance more strongly than glycemia) and provide fecal samples, the latter of which were subjected to fecal microbial profiling via metagenomics assays.
Study Findings
Study participants were observed to be between 50 and 64 years old, with OGTT-based subgroup classification revealing 220 participants with NGT, 185 with IFG, 173 with IGT, 74 with CGI, and 45 with screen-detected T2D. Participants’ blood plasma metabolomics revealed 978 plasma metabolites obtained primarily from the metabolism of lipids (45.4%) and amino acids (22.1%).
Gradient-boosted decision tree (GBDT) models revealed 645 metabolites in the discovery cohort significantly associated with IFG, IGT, CGI, or T2D. Of these, 502 metabolites overlapped in significance in the validation cohort, suggesting their role as potential biomarkers of glucose control (prediabetes and T2D biomarkers). Notably, 143 of these metabolites were associated with microbiome data and 272 with diet data.
“These findings show that potential determinants persist in prediabetes and T2D, with the gut microbiome alone accounting for nearly one-third of blood metabolite variance, twice that measured in healthy individuals.”
Identified metabolomics profiles were found to overlap with previously identified signatures of prediabetes, T2D, acute coronary syndrome (ACS), heart failure (HF), and kidney disease (KD). This confirms that microbiome-metabolome dynamics are perturbed before the onset of CVD, thereby suggesting potential early intervention targets against cardiometabolic disease incidence. For example, the metabolite hippurate mediated interactions between specific gut bacteria (Hominifimenecus microfluidus and Blautia wexlerae), with bidirectional mediation effects observed (21.1% of H. microfluidus’s influence on B. wexlerae was mediated by hippurate).
Co-analysis of lifestyle and metabolome data revealed that ~65.9% of identified metabolite biomarkers are associated with reversible lifestyle changes, highlighting the potential for monitoring the effects of exercise or diet interventions in successfully preventing or treating diabetic outcomes. High coffee intake, common in the Swedish cohort, reduced diet-related metabolite variability, underscoring population-specific microbiome adaptations. Notably, the metabolite imidazole propionate was elevated in IGT but not validated in the SCAPIS cohort, suggesting population-specific variability.
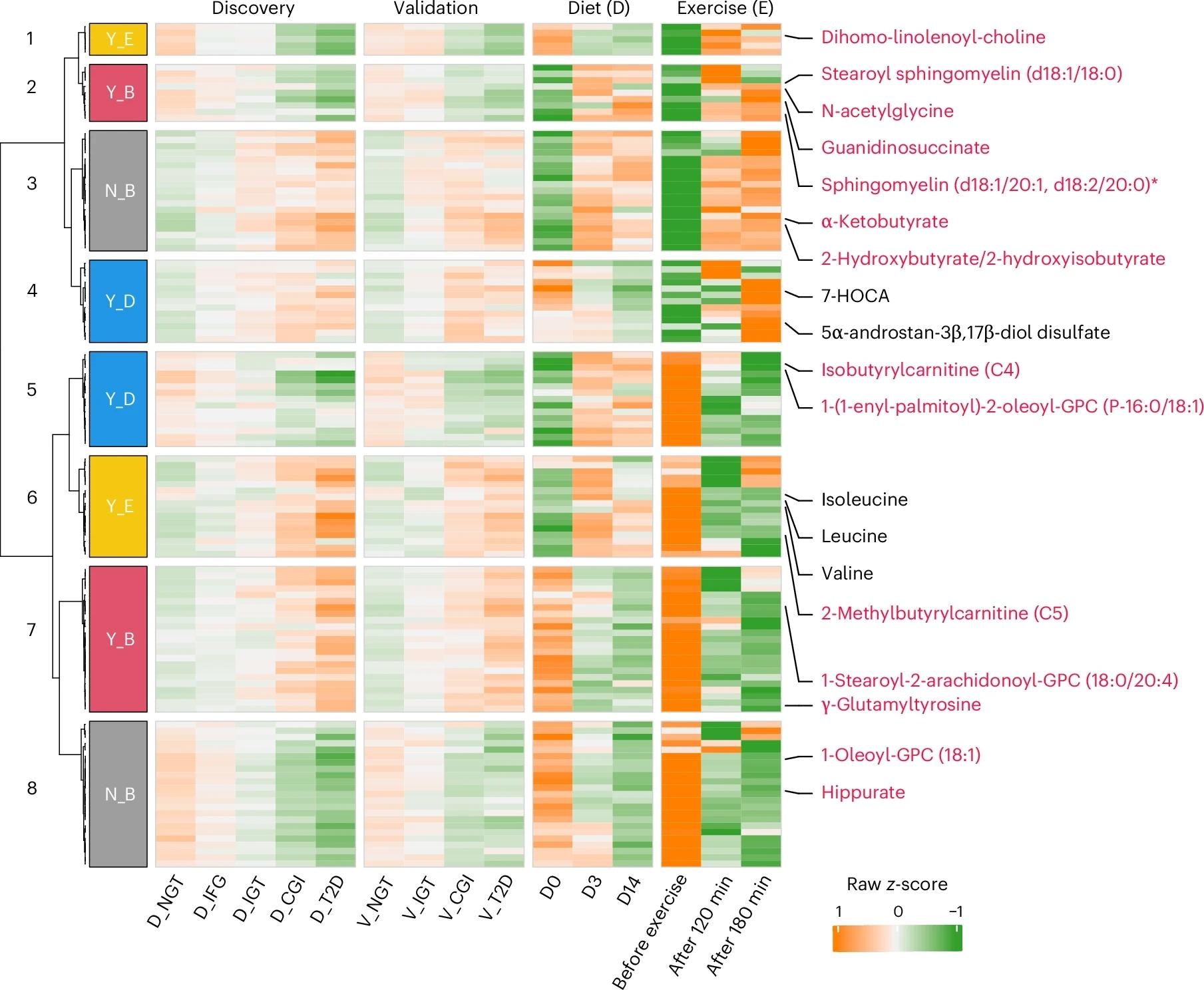 Heatmap showing the overlapping metabolites involved in amino acid, lipid and xenobiotic metabolism (n = 123) in two clinical trials of either diet (14 days) or exercise for 1-h (before, 120 and 180 min after exercise) interventions with those 502 altered metabolites in prediabetes and T2D. Responses reversed (Y, yes; N, no) by either diet (D) or exercise (E) or both (B) were clustered and are shown in distinct colors beside the row clustering branches. Representative metabolites, including 14 overlapping with Fig. 4f, are labeled in red, and five others in black. Wilcoxon rank-sum test and one-way repeated-measures analysis of variance were used to identify altered metabolites in the cohorts and two longitudinal datasets (Padj < 0.1), respectively.
Heatmap showing the overlapping metabolites involved in amino acid, lipid and xenobiotic metabolism (n = 123) in two clinical trials of either diet (14 days) or exercise for 1-h (before, 120 and 180 min after exercise) interventions with those 502 altered metabolites in prediabetes and T2D. Responses reversed (Y, yes; N, no) by either diet (D) or exercise (E) or both (B) were clustered and are shown in distinct colors beside the row clustering branches. Representative metabolites, including 14 overlapping with Fig. 4f, are labeled in red, and five others in black. Wilcoxon rank-sum test and one-way repeated-measures analysis of variance were used to identify altered metabolites in the cohorts and two longitudinal datasets (Padj < 0.1), respectively.
Conclusions and Future Directions
The present study reveals the role of microbiome-metabolome dynamics in altering human glucose homeostasis, triggering prediabetes and T2D. It highlights the importance and potential of lifestyle changes, particularly diet and exercise, in adjusting and monitoring these dynamics to achieve optimal health outcomes. The findings were further validated in GF/CONV-R mice and external cohorts (Israeli, TwinsUK), strengthening their robustness. Optimal benefits likely require combining diet and exercise interventions, as demonstrated by lifestyle-specific metabolite modulation (e.g., branched-chain fatty acids improved with exercise, 7-HOCA with diet).
“Understanding the connections between diet, gut microbiota, and clinical factors provides valuable insights into T2D and highlights the need for diverse intervention strategies. This resource may provide increased understanding of how gut microbiota may affect T2D and help identify new targets for diabetes management.”
The study authors have developed an open-access web server (https://omicsdata.org/Apps/IGT_metabolome/), which will provide future researchers with an easy-to-use platform for metabolome exploration, meta-analysis, and data visualization.