At the end of 2019, Chinese officials claimed that SARS-CoV-2 infections were only identified in patients who previously attended the Huanan Seafood Market, thus indicating that human-to-human transmission of the virus was not occurring. However, by January 2020, it became clear that SARS-CoV-2 was spreading between humans, with some of the earliest coronavirus disease 2019 (COVID-19) cases having no relation to the Huanan Seafood Market.
Before the Huanan Seafood Market was closed on January 1, 2020, about one month after the first COVID-19 case was detected in China, the Chinese CDC collected samples from 457 animal species of the Huanan Seafood Market, none of which tested positive for SARS-CoV-2. Comparatively, 73 of the 923 environmental samples from the same market were positive for SARS-CoV-2.
This data, which was eventually published in a 2022 preprint, only correlated the number of SARS-CoV-2 deep sequencing reads with the number of human reads. In addition to not providing any specific correlations for other species, the Chinese scientists also did not provide the raw sequencing data, which could have been used by other researchers around the world to determine whether the abundance of SARS-CoV-2 genetic material identified in these samples could be correlated with material from other species.
Nevertheless, the Chinese CDC eventually uploaded the raw sequencing data for some of the environmental samples to the Global Initiative on Sharing All Influenza Data (GISAID) database. Thereafter, bioinformatic analysis of this data led to the identification that some of the environmental samples contained genetic material from animals like raccoon dogs that are susceptible to SARS-CoV-2.
Study findings
In an effort to provide an objective analysis of these environmental samples, the author of the current study devised a fully reproducible computational pipeline using data originally published by the Chinese CDC.
The dataset, which consisted of 696 FASTQ files from 365 deep sequencing runs of 176 samples, was processed to retain only high-quality leads. Dr. Bloom subsequently aligned each read to the SARS-CoV-2 genome and a representative set of mammalian mitochondrial genomes. This analysis similarly reported that the mitochondrial genetic material of raccoon dogs was most abundant, followed by that of ducks.
When the author’s computational pipeline was applied to the environmental samples, over 75% of these samples did not exhibit any reads that aligned to the SARS-CoV-2 genetic material, with most of the remaining 25% also exhibiting only a small number of viral reads. In fact, only two samples had over 1,000 reads that aligned with SARS-CoV-2. Moreover, most of the samples with high SARS-CoV-2 content were collected by the Chinese CDC on January 1, 2020, with few SARS-CoV-2-positive samples collected after this date.
Dr. Bloom also analyzed any correlations that existed between the number of reads mapped to SARS-CoV-2 to the mitochondrial genome of different species identified across all samples. When Dr. Bloom applied a log-log scale to this data, which was also performed in the original Chinese CDC report, the genetic material of the largemouth bass, catfish, cow, carp, and snakehead fish were most correlated with SARS-CoV-2 abundance. Importantly, none of these animals are likely hosts for SARS-CoV-2.
Due to conflicting results regarding correlations between human and SARS-CoV-2 genetic material from the collected samples, the author used chordate mitochondrial compositions. To this end, when only samples containing at least one SARS-CoV-2 read were used, the genetic material of the largemouth bass, catfish, cow, sheep, and pig were all more strongly correlated with SARS-CoV-2 reads than that of humans.
Although the degree of correlation increased between SARS-CoV-2 and human genetic material when samples collected before January 12, 2020, were excluded, Dr. Bloom found that the genetic material of goats and spotted doves were also similarly correlated. Notably, neither analysis indicated that the genetic material of raccoon dogs nor bamboo rats was positively correlated with SARS-CoV-2 reads.
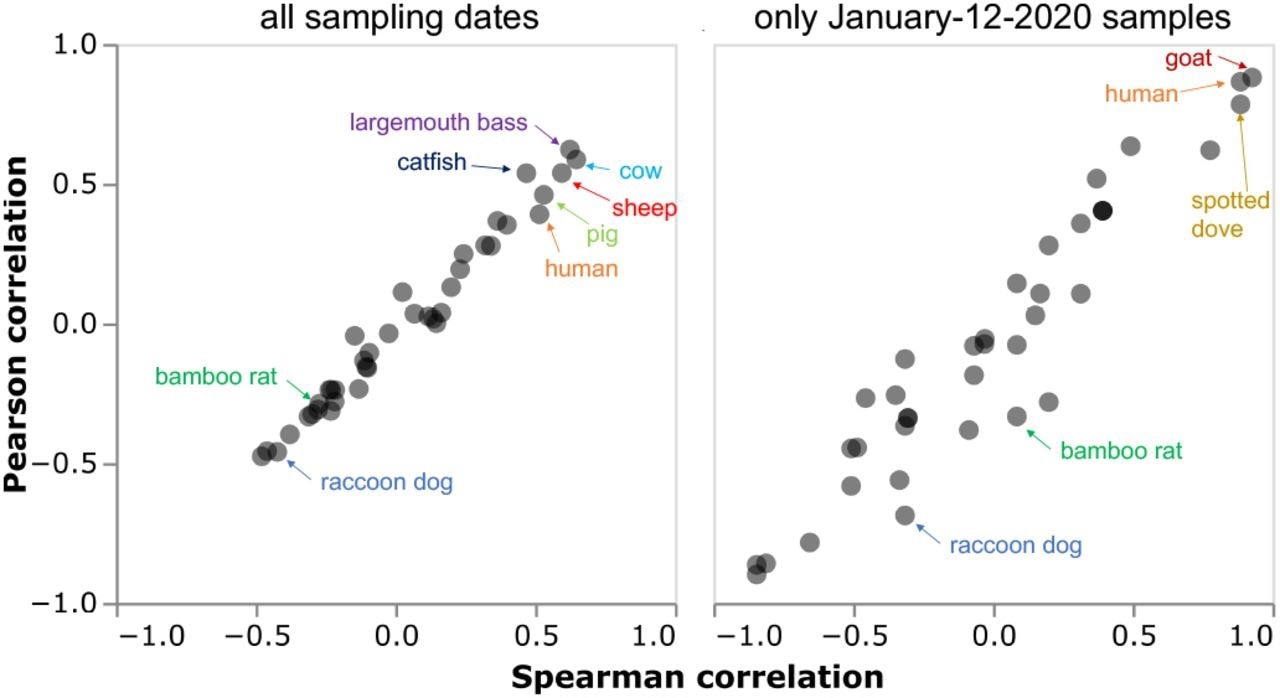
Correlations between SARS-CoV-2 content and mitochondrial content for all species were calculated for samples containing at least one SARS-CoV-2 read from any sampling date (left) or just the January-12-2020 date when most of the wildlife sampling occurred. This plot is designed to mimic the fourth figure of Liu et al. (2022), and so only shows samples with at least one SARS-CoV-2 read and calculates correlations between the log number of SARS-CoV-2 reads versus the log number of species mitochondrial reads for consistency with Liu et al. (2022). See the interactive version at https://jbloom.github.io/Huanan_market_samples/overall_corr.html to mouseover all points for details, select different subsets of samples, and calculate the correlations on a linear or log scale.
Conclusions
The author of the current study confirmed that many of the environmental samples collected from the Huanan Seafood Market in January 2020 contained genetic material from various species, including humans, fish, snakes, cows, goats, pigs, sheep, birds, raccoon dogs, and bamboo rats. After expanding his analysis to correlate the abundance of SARS-CoV-2 and mitochondrial genetic material across all environmental samples, Dr. Bloom observed that the greatest co-mingling of viral and animal material involved species that were almost certainly not infected by SARS-CoV-2, such as fish and livestock.
There was a weak but identifiable correlation between the abundance of SARS-CoV-2 and human genetic material. Furthermore, the genetic material of raccoon dogs was extremely unlike to correlate to SARS-CoV-2 genetic material.
Most of the samples analyzed in this study were collected on January 1, 2020, or later, which is several weeks after the Huanan Seafood Market acted as a superspreading site for human SARS-CoV-2 infections. Thus, it is not surprising that the analysis of these samples did not elucidate the exact origin of SARS-CoV-2.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Bloom, J. D. (2023). Association between SARS-CoV-2 and metagenomic content of samples from the Huanan Seafood Market. bioRxiv. doi:10.1101/2023.04.25.538336