The intensive research during the coronavirus disease 2019 (COVID-19) pandemic has helped understand SARS-CoV-2 infection. Nevertheless, what makes the virus capable of causing a dangerous inflammatory response remains unclear. Research has suggested that amphiphilic, cationic peptides from the innate immune system undergo amyloid-like assembly with anionic nucleic acids and form proinflammatory complexes.
 Study: Viral afterlife: SARS-CoV-2 as a reservoir of immunomimetic peptides that reassemble into proinflammatory supramolecular complexes. Image Credit: NIAID
Study: Viral afterlife: SARS-CoV-2 as a reservoir of immunomimetic peptides that reassemble into proinflammatory supramolecular complexes. Image Credit: NIAID
The study and findings
The present study investigated whether fragmented SARS-CoV-2 peptides assemble with anionic double-stranded RNA (dsRNA) into supramolecular complexes. The viral proteome was considered a reservoir of peptide fragments liberating after the proteolytic destruction of virions. The researchers leveraged a support vector machine (SVM) classifier to recognize antimicrobial peptide (AMP)-like sequences (xenoAMPs) in the SARS-CoV-2 proteome.
Viral protein sequences were scanned via a moving window of 24–34 amino acids to identify potential xenoAMPs and test whether they behave like AMPs if cleaved at different positions. Sequences were selected based on the output provided by the classifier as a sigma (σ) score, wherein a strongly positive score implied the sequence was highly likely to be an AMP.
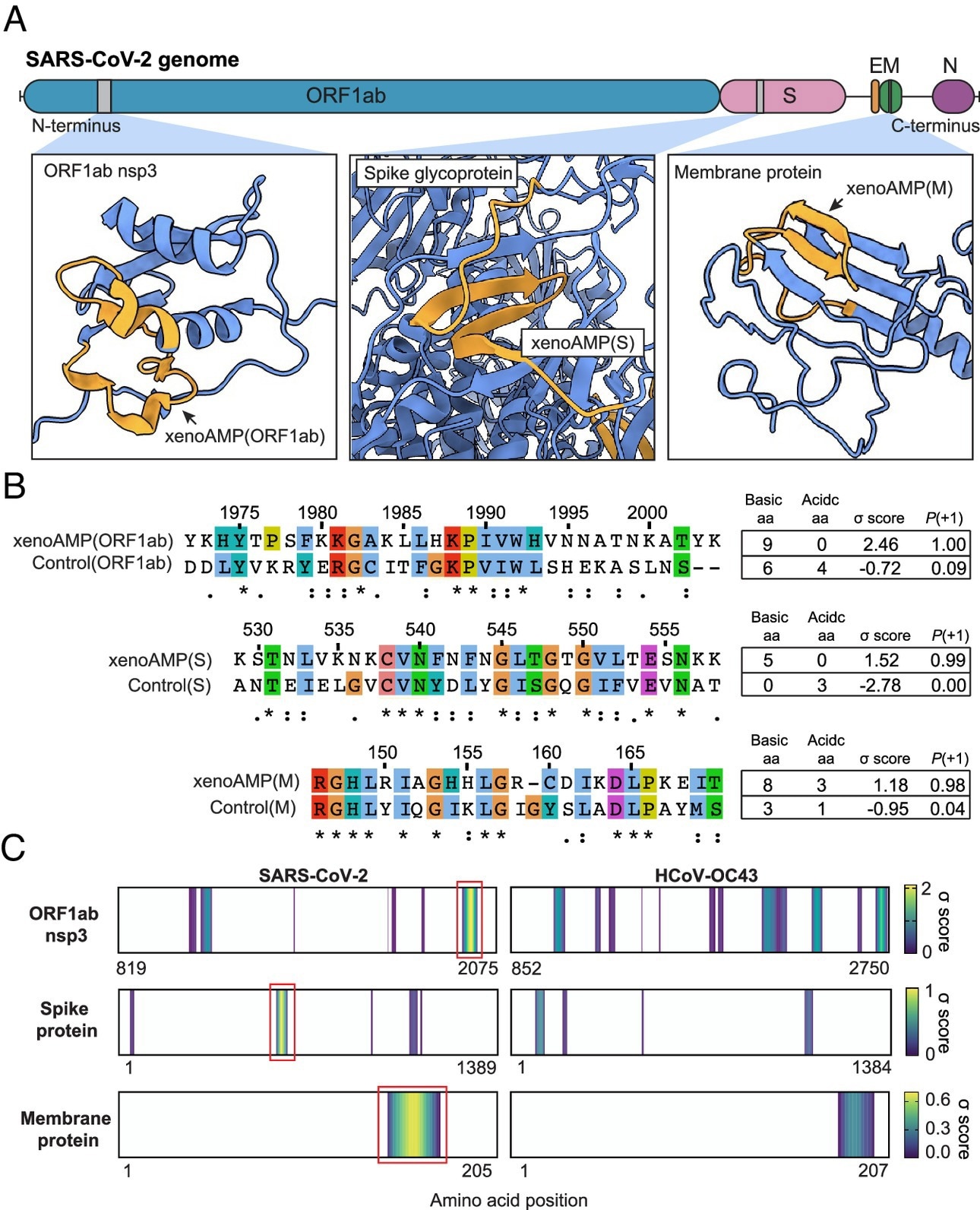
Existence of exogenous mimics of pro-inflammatory host antimicrobial peptides (xenoAMPs) in SARS-CoV-2 proteins. (A) SARS-CoV-2 proteins are scanned with a machine-learning AMP classifier. Each queried sequence is given a σ score that measures its AMP-ness. Three representative high-scoring sequences are studied: xenoAMP(ORF1ab), xenoAMP(S), and xenoAMP(M). The grey bars mark the location where the corresponding sequences are selected. (B) SARS-CoV-2 sequences are aligned and compared to their homologs in a common cold human coronavirus HCoV-OC43: Control (ORF1ab), Control(S), and Control(M). Asterisks, colons, and periods indicate positions that have fully conserved residues, those that have strongly similar properties, and those that have weakly similar properties, respectively. Color is assigned to each residue using the ClustalX scheme. (C) σ score heatmaps compare the distribution of high-scoring sequences in three proteins from SARS-CoV-2 and HCoV-OC43. The first amino acid in each sequence is colored according to its average σ score; regions with negative average σ scores (non-AMPs) are colored white. “Hot spot” clusters of high-scoring sequences for SARS-CoV-2 (bright yellow regions bracketed in red boxes) have systematically higher scores and span wider regions of sequence space compared to HCoV-OC43. This trend suggests that hot spots in SARS-CoV-2 can generate higher scoring sequences for a greater diversity of enzymatic cleavage sites than those in HCoV-OC43.
Further, the team selected specific sequences from this population of (high-scoring) sequences with a high cationic charge. Specifically, they focused on prototypical candidates from the membrane (M) protein, spike (S) protein, and open reading frame 1ab (ORF1ab) polyprotein. In silico analyses showed that these xenoAMPs could be generated during proteasomal degradation, with matrix metalloproteinase 9 (MMP9) and neutrophil elastase (NE) capable of generating them.
Next, the team compared SARS-CoV-2 xenoAMPs with homologous sequences from SARS-CoV-1 and non-pandemic human CoVs. This showed that sequences were partially conserved. A comparison of σ score heat maps of ORF1ab, S, and M proteins between SARS-CoV-2 and HCoV-OC43 revealed that high-scoring sequences were clustered into hotspots, with SARS-CoV-2 hotspots having higher scores and spanning wider regions than those of HCoV-OC43.
Further, mass spectrometry was performed on tracheal aspirate samples from patients with severe COVID-19. The team detected fragments of host AMP, cathelicidin LL-37, in 20 samples (out of 29). By contrast, 28 samples contained viral peptide fragments, some of which had sufficiently high σ scores to qualify as xenoAMPs.
The three xenoAMPs, xenoAMP(S), xenoAMP(M), and xenoAMP(ORF1ab), were experimentally observed to chaperone and assemble with dsRNA into complexes similar to LL-37. Polyinosine: polycytidylic acid (Poly(I:C) was used as a synthetic analog to mimic the viral dsRNA generated during replication. The structures of xenoAMPs-poly(I:C) complexes were cognate to host AMPs-dsRNA complexes.
Next, the team investigated the robustness of these self-assembled proinflammatory complexes under non-optimal conditions. They found that the nanocrystalline structures were preserved when participating xenoAMPs were shortened. Besides, SARS-CoV-2 xenoAMPs were found to co-crystallize with LL-37, suggesting that host AMPs and xenoAMPs could synergistically activate inflammatory responses.
The immune activation capacity of xenoAMPs from SARS-CoV-2 was compared with that of homolog peptides from HCoV-OC43 using human monocytes. XenoAMP-poly(I:C)-treated monocytes released 1.7-fold more interleukin (IL)-8 than poly(I:C) treated controls. By contrast, complexes formed with homologous peptides from HCoV-OC43 induced much lower IL-8 levels.
In addition, xenoAMP-poly(I:C) stimulation of primary human dermal microvascular endothelial cells (HDMVECs) triggered robust production of IL-6, which was not observed with complexes formed from HCoV-OC43 peptides. Notably, xenoAMP-poly(I:C)-treated HDMVECs showed significant upregulation of several proinflammatory chemokine and cytokine genes.
Finally, the researchers measured the immune activation capacity in mice. C57BL/6 mice unexposed to infection were treated with xenoAMP(ORF1ab)-poly(I:C) complexes or poly(I:C)-alone (control). XenoAMP(ORF1ab)-poly(I:C) treatment increased plasma levels of IL-6 and C-X-C motif chemokine ligand 1 (CXCL1) by 1.6 and 2.2 times, respectively, compared to poly(I:C)-alone. Moreover, IL-6 and CXCL1 levels increased 1.2 times in the lung compared to the control treatment.
Conclusions
In sum, the study has illustrated an unexpected mechanism of inflammation propagating through uninfected cells in COVID-19, wherein viral fragments mimic AMPs like LL-37. This could be salient to understand why the host immune system in COVID-19 resembles that of individuals with autoimmune conditions like rheumatoid arthritis and lupus.
The researchers found that host proteases could generate xenoAMPs, suggesting that protease inhibitors suppressing xenoAMP generation could have a clinical impact on viral-induced inflammation. The proteolytic degradation of SARS-CoV-2 could differ across host individuals, possibly explaining the heterogeneity of infection outcomes, e.g., asymptomatic and fatal.
Journal reference:
- Zhang Y, Bharathi V, Dokoshi T, et al. Viral afterlife: SARS-CoV-2 as a reservoir of immunomimetic peptides that reassemble into proinflammatory supramolecular complexes. Proc Natl Acad Sci USA, 2024, DOI: 10.1073/pnas.2300644120, https://www.pnas.org/doi/10.1073/pnas.2300644120