Domain generalization has become a primary issue for ML use in healthcare settings since model performance may be worse than planned due to data discrepancies during model development and deployment. Underrepresentation of specific groups or diseases is a typical problem that competent doctors struggle to solve due to disease rarity or the availability of clinical knowledge. Few initiatives have gained widespread acceptance and scaled influence on clinical outcomes, with 'out-of-distribution' data being a significant hurdle to implementation.
About the study
In the present study, researchers employed diffusion models to examine medical imaging situations such as histology, chest X-rays, and dermatological pictures. They used these photos to enhance the reliability and fairness of medical machine-learning models. They also utilized unlabeled data to track data dispersion and supplement actual samples. The project sought to expand the training dataset in a steerable and programmable manner.
The researchers trained a generative model using labeled and unlabeled data, with labeled data accessible exclusively for a single source domain and extra unlabeled data from any domain (in or out of distribution). They may condition the model on diagnostic labels with or without property (for instance, sensitive attribute labels or ID of hospitals). The researchers de-identified data before analysis. Model conditioning on one or both qualities allowed them to specify which synthetic samples were used to supplement the training set. They trained the generative model of low resolution and upsampler using one conditioning vector.
The team added synthetic pictures from generative modeling to training data obtained from source domains before diagnostic model training. They tested their strategy on several medical situations with denoising diffusion probabilistic models (DDPMs), tracking fairness and diagnostic performance in and out from distribution (OOD). They defined in-distribution data as photos from similar demographic and illness distributions obtained using the one imaging technique as training data.
The researchers used two criteria to compare the model baseline performances and the suggested technique. One set concentrated on diagnostic accuracies, such as top-1-type accuracy in identifying histology images and receiver operating curve-area under the curve (ROC-AUC) values for radiological assessments, whereas the other was more concerned with fairness. Expert dermatologists have found high-risk-type sensitivity to be the most useful diagnostic tool.
Researchers used two big public radiology datasets, CheXpert and ChestX-ray, to create generative and diagnostic models for chest X-rays. After training on 201,055 chest X-ray instances, dermatologists evaluated the model's ability to capture primary characteristics on 488 synthetic pictures from regular and high-risk classes. They assessed the picture quality to offer a diagnosis for up to three of the approximately 20,000 common illnesses.
Results
The study shows that diffusion models may learn realistic augmentations from data in a label-efficient way, making them more resilient and statistically fair both in and out of distribution. Combining synthetic and real-time data can considerably increase diagnostic accuracy and decrease the fairness gap between different qualities during shifts in distribution.
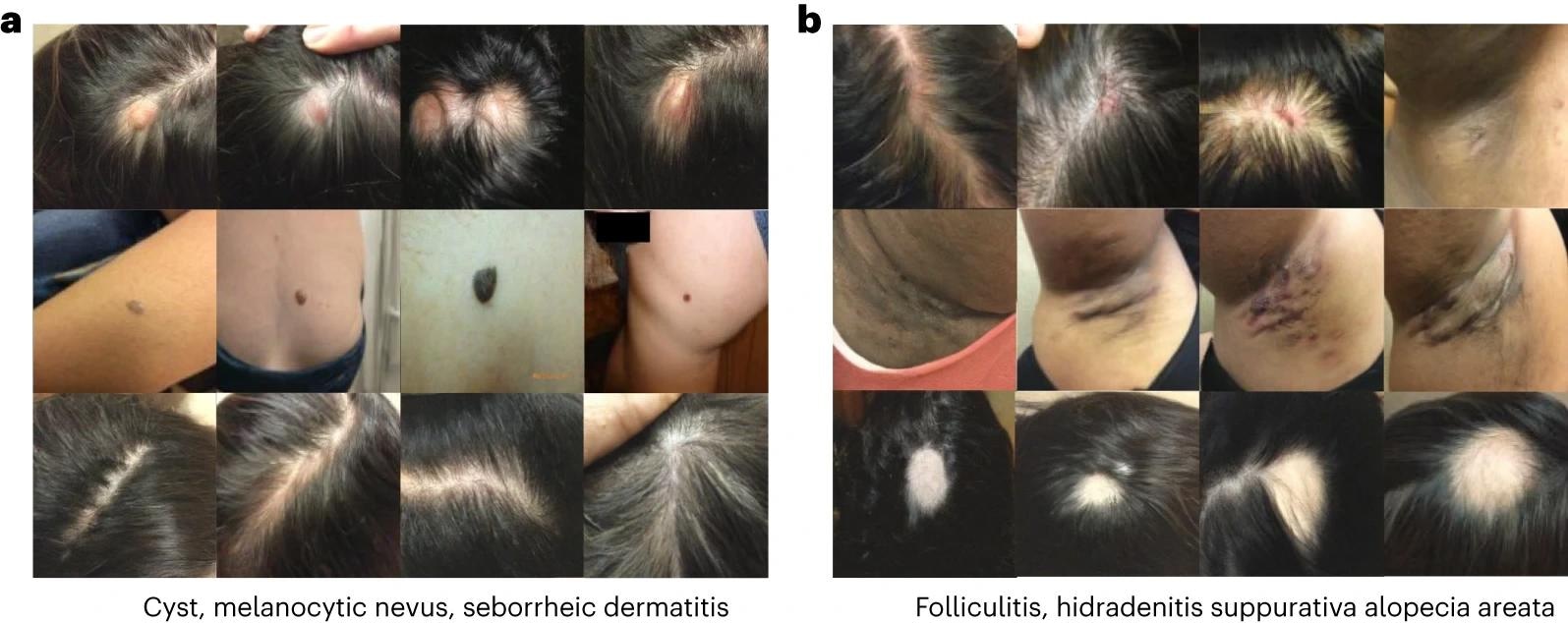 Generated images in the dermatology setting. Each row of images corresponds to a different condition. a, Generated images for cyst, melanocytic nevus and seborrheic dermatitis. b, Generated images for folliculitis, hidradenitis and alopecia areata.
Generated images in the dermatology setting. Each row of images corresponds to a different condition. a, Generated images for cyst, melanocytic nevus and seborrheic dermatitis. b, Generated images for folliculitis, hidradenitis and alopecia areata.
While not a substitute for representative and high-quality data collection methods, it can enable clinicians to use unlabeled and labeled information and close potentially harmful diagnostic accuracy gaps between underrepresented and overrepresented populations without penalizations. The researchers found that using synthetic data beat in-distribution baselines in more and less skewed circumstances, narrowing fairness gaps between hospitals.
Color augmentations on top of produced samples performed the best overall, with 49% relative improvements over baseline modeling and a 3.2% improvement over models with color augmentation training in the test hospital. The study showed that synthetic pictures considerably increased the average AUC for five diseases, notably cardiomegaly and OOD. The female fairness difference narrowed by 45%, while the race fairness gap shrank by 32%. Combining heuristic augmentations with synthetic data-based techniques such as 'Label conditioning' and 'Label and property conditioning' increased model sensitivity without sacrificing fairness, resulting in considerable gains in OOD scenarios.
Label and property conditioning improved high-risk diagnostic sensitivity by 27% and increased OOD by 63.5%, narrowing the fairness gap by 7.5×. The dermatological modality produced realistic and canonical pictures that captured features of numerous illnesses, including rare occurrences. Synthetic pictures also decreased false correlations and compressed representations, lowering the model's dependence on non-generalizable OOD correlations and underserving individuals.
The study shows that diffusion models may generate synthetic pictures helpful in medical applications such as histology, radiology, and dermatology while enhancing statistical fairness, balanced accuracy, and high-risk sensitivity. These synthetic samples produce realistic, canonical pictures that professional doctors consider diagnosable. However, the researchers point out possible hazards and limits depending on created data, such as overconfidence in AI systems, restricted insights, and the recurrence of biases in the original training data.