Inflammatory and autoimmune disorders affect about 5% of the global population and are highly heterogeneous, affecting various parts of the body. There is a dearth of effective therapies to address inflammatory diseases such as inflammatory bowel disease, lupus, and psoriasis, with less than 10% of the drugs undergoing clinical testing being approved for use. An inadequate understanding of the mechanisms of inflammatory diseases is the primary cause of the low efficacy of inflammatory disease therapies.
However, advancements in genetics and the mapping of numerous loci present an avenue to examine the loci directly linked to disease pathogenesis. Identifying the pathways implicated by these genetic loci presents opportunities to develop drugs specifically targeting these pathways.
About the study
In the present study, the researchers investigated shared disease mechanisms and aimed to identify genetic variants that determined the predisposition to multiple inflammatory diseases. They identified the intergenic region present on the 22nd position of the long arm (q) of the 21st chromosome (chr21q22), which contains the haplotype determining the predisposition to five inflammatory diseases.
They conducted a co-localization analysis to test whether the disease mechanism was common across the five diseases. They hypothesized that since all five inflammatory diseases were immune-mediated, the chr21q22 locus was involved in immune function.
The study used chromatin immunoprecipitation-sequencing to identify the active promoters and enhancers based on the hypothesis that the chr21q22 locus had a distal enhancer function in immune mechanisms.
Using a combination of the human monocyte’s expression quantitative locus data and promoter-capture Hi-C, the researchers then scanned numerous genes in close proximity to the chr21q22 locus to identify which gene was regulated by the enhancer at chr21q22.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system was then used to confirm the causal role of the identified gene. For this, the researchers selectively deleted the enhancer region in human monocytes using CRISPR/Cas9 and then cultured these altered monocytes with inflammatory ligands such as prostaglandin E2 and tumor necrosis factor, which are proinflammatory lipids and cytokines, respectively, and a toll-like receptor agonist Pam3CSK4.
The expression levels of messenger ribonucleic acid (mRNA) of all the candidate genes were then measured to determine which gene was affected by the deletion of the enhancer at chr21q22. Data from a genome-wide association study on inflammatory bowel disease were then used to determine the causal variant of the enhancer.
The high levels of linkage disequilibrium between selected single nucleotide polymorphisms made it challenging to identify the causal variant of the enhancer. Therefore, massively parallel reporter assay helped screen numerous short deoxyribonucleic acid (DNA) sequences to test for enhancer activity.
Results
The study found that the intergenic region on chr21q22 enhanced the Erythroblast Transformation Specific proto-oncogene 2 (ETS2), which centrally regulates inflammatory macrophages in humans.
Diseased tissues were found to prominently express the genes regulated by the ETS2 protein. These genes were also more enriched in the genome-wide association study data for inflammatory bowel disease than other pathways.
Furthermore, when ETS2 was overexpressed in macrophages, the inflammatory state that is observed in the five inflammatory diseases linked to the chr21q22 locus was re-established, and various drug targets, such as interleukin 23 and tumor necrosis factor, were upregulated.
The researchers also aimed to identify potential treatment methods for inflammatory diseases. One of these was to use mitogen-activated protein kinase kinase or MEK inhibitors to block ETS2 signaling, which can target multiple cytokines. However, this method can have numerous side effects because MEK also plays various roles in other tissues.
Therefore, the researchers believe that using proteolysis-targeting chimera molecules or PROTACs to target the ETS2 protein directly or selectively delivering MEK inhibitors to macrophages using antibody-drug conjugates can circumvent the side effects and successfully block the ETS2-based inflammation.
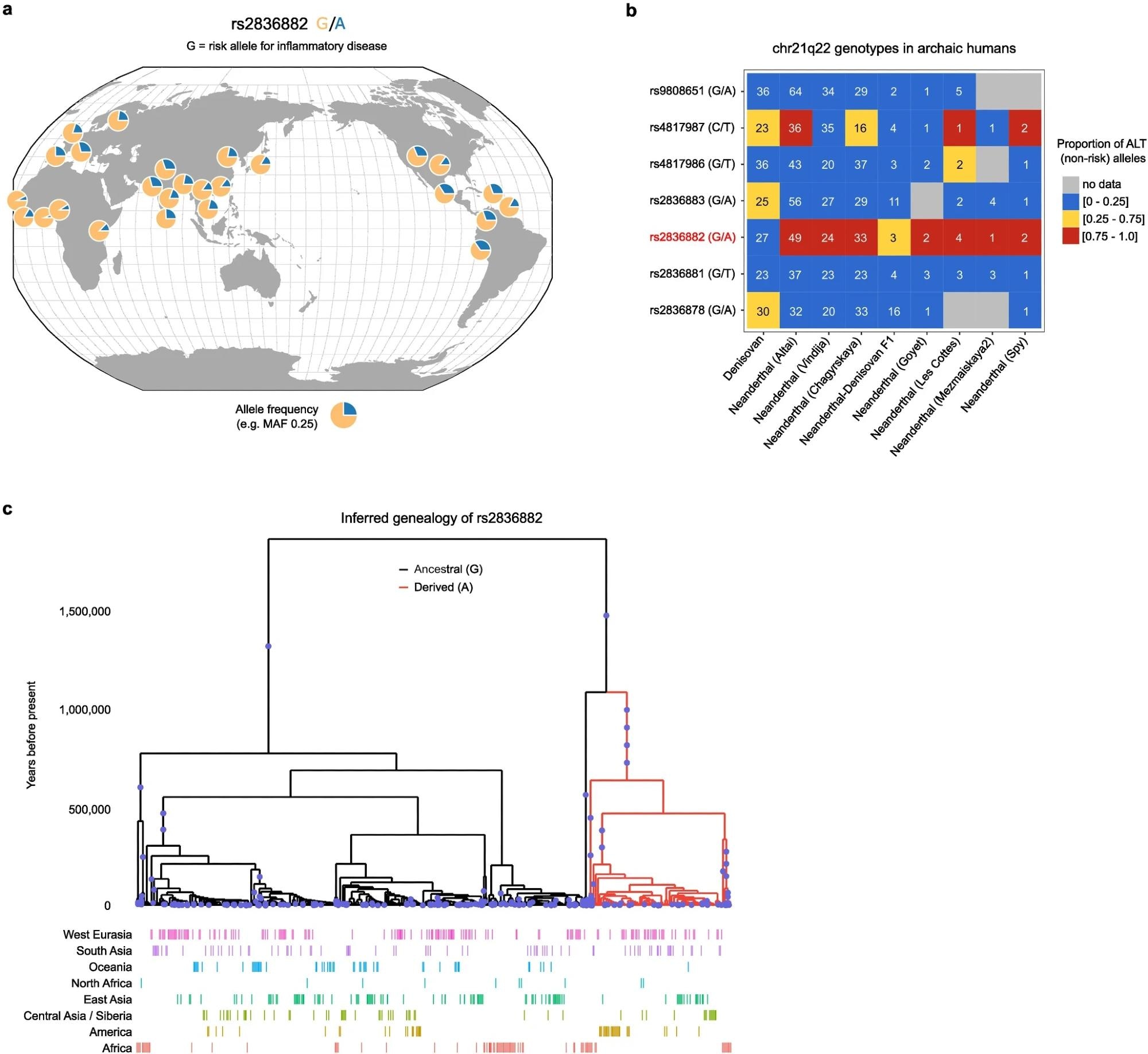
Geographic distribution and history of rs2836882. a. rs2836882 allele frequency in modern global populations (data from 1000 Genomes Project, plotted using Geography of Genetic Variants browser: https://popgen.uchicago.edu/ggv/). b. Genotypes of candidate SNPs at chr21q22 (99% credible set) in archaic humans (Neanderthals and Denisovans). Colour depicts the proportion of reads containing ALT alleles, with a value close to 0 consistent with a homozygous REF (risk) genotype, a value close to 1 consistent with a homozygous ALT (non-risk) genotype, and an intermediate value indicating a potential heterozygous genotype. Number in each cell indicates the number of reads at that SNP in the indicated sample. Putative causal variant highlighted in red. c. Inferred genealogy of the age of the rs2836882 polymorphism – analysed using Relate. The diagram in a was created using the Geography of Genetic Variants browser.
Conclusions
Overall, the findings reported that the ETS2 gene was enhanced by the intergenic region on chr21q22, which is implicated in five inflammatory diseases. Methods to directly target the ETS2 protein or selectively inhibit ETS2 signaling in macrophages could be potential treatment methods for inflammatory diseases.