The development of clinically useful tools to identify individuals at an increased risk of disease remains a significant challenge in precision medicine. The lack of these tools often delays diagnoses, thereby contributing to adverse patient outcomes.
Single plasma proteins can be used to diagnose certain diseases, such as troponins for acute coronary syndromes. Plasma proteomic signatures can also reflect current health status, health behaviors, and even genetic and environmental determinants of disease.
It remains unclear whether plasma proteomics can be used to predict certain diseases alone or in combination with other markers. Previous studies examining the role of plasma proteomics in diagnostics have been associated with certain limitations, including small sample sizes and the inclusion of a few common diseases, rather than taking an agnostic discovery approach. There is also a lack of evidence on the accuracy of screening metrics compared to clinical models without proteins.
About the study
The study sample comprised 41,931 individuals from the United Kingdom Biobank Pharma Proteomics Project. The measurements of about 3,000 plasma proteins were integrated with clinical information to construct sparse prediction models for 218 rare and common diseases. These diseases were associated with high morbidity and/or mortality rates.
To construct the model, 80 incident cases within 10 years of follow-up were isolated from the random U.K. Biobank. Incident cases contained in the ‘consortium-selected’ subset were also used.
The integration of data from primary care, cancer and death registries, hospital episode statistics, and self-reported illnesses led to validated phenotypes on which disease definitions were based. Incident cases registered within the first six months of follow-up were excluded from the study. Prevalent cases or those recorded before the baseline assessment visit were also excluded.
Proteomic profiling captured 2,923 unique proteins targeted by 2,941 assays. A three-step machine learning (ML) framework was adapted, which included selecting features, tuning, optimizing hyperparameters, and validating.
Half of the sample was used for feature selection, whereas the other half was equally subdivided for model optimization and validation. The proteomic data-based models were then compared with those based on either clinical information alone or a combination of clinical information and data from 37 clinical assays.
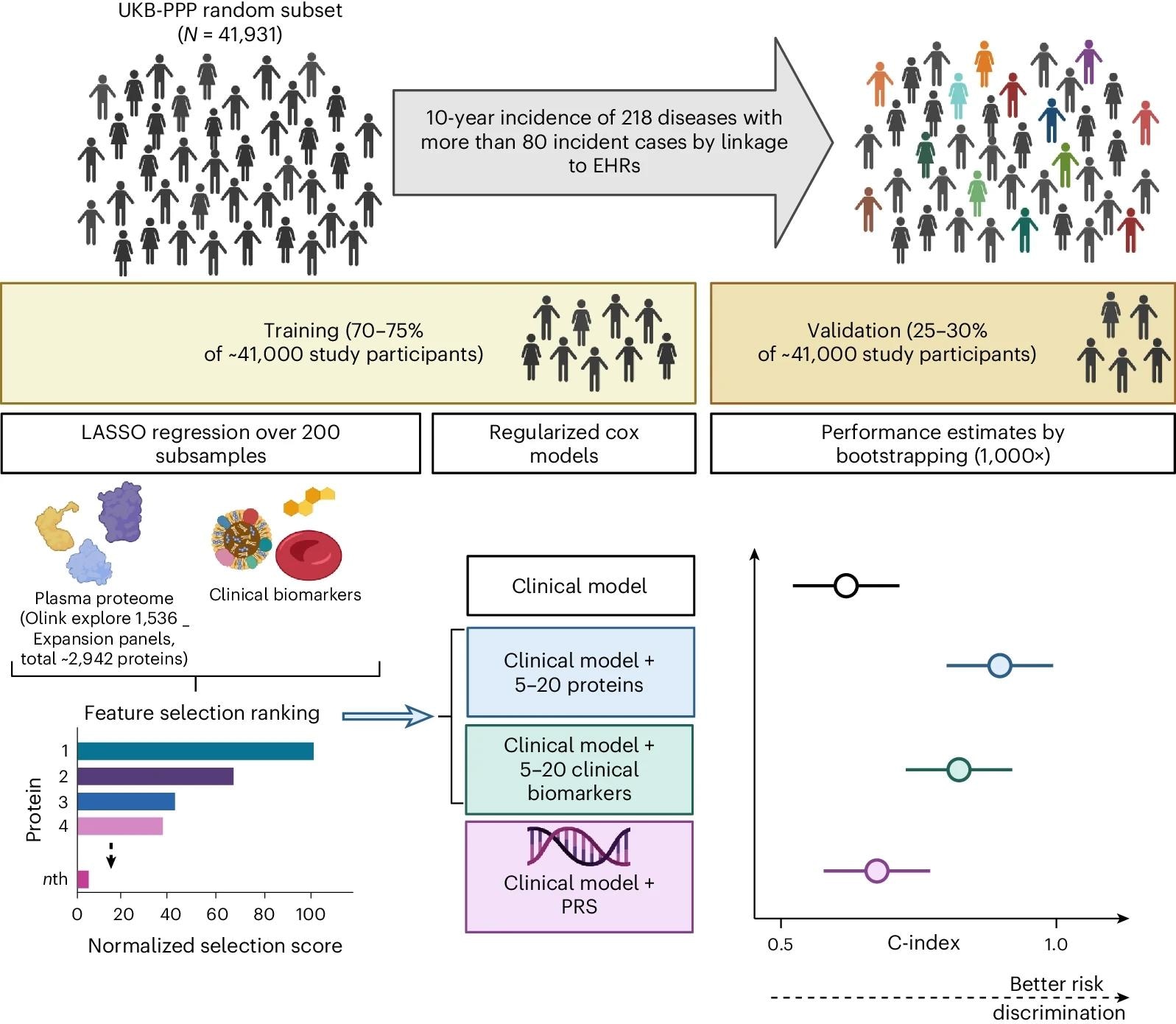 This cohort study is based on a random subset of UKB-PPP individuals (N = 41,931). The cohort was divided into training (including feature selection and optimization steps) and validation sets to develop sparse protein-based predictors (including 5–20 proteins from the Olink Explore 1536 and Explore Expansion panels) for 218 diseases defined using data from the UKB health-questionnaire, primary care, hospital episode statistics and cancer and death registries. Performance of models using protein signatures was compared with models using basic clinical information alone or using basic clinical information combined with clinical assay data or genome-wide PGS. Created with BioRender.com.
This cohort study is based on a random subset of UKB-PPP individuals (N = 41,931). The cohort was divided into training (including feature selection and optimization steps) and validation sets to develop sparse protein-based predictors (including 5–20 proteins from the Olink Explore 1536 and Explore Expansion panels) for 218 diseases defined using data from the UKB health-questionnaire, primary care, hospital episode statistics and cancer and death registries. Performance of models using protein signatures was compared with models using basic clinical information alone or using basic clinical information combined with clinical assay data or genome-wide PGS. Created with BioRender.com.
Study findings
Clinical models achieved the highest performance for endocrine and cardiovascular diseases. For a subset of 163 diseases, five proteins alone performed as well as the clinical model and significantly better for an additional 30 diseases.
Incorporating five to 20 proteins significantly improved the performance of clinical models for 67 rare and common diseases, including celiac disease, motor neuron disease, and pulmonary fibrosis. For 28 diseases, models incorporating blood assay data outperformed clinical models.
For 52 of these 67 diseases, models with sparse protein signatures outperformed clinical models with blood assays. Including proteins augmented the models specifically for less common diseases.
The strong predictive power of proteins was demonstrated for newly diagnosed multiple myeloma (MM) patients. Single-cell ribonucleic acid (RNA) sequencing from bone marrow highlighted that four of the five predictor proteins, including Fc receptor-like B (FCRLB), glutaminyl-peptide cyclotransferase (QPCT), SLAM family member 7 (SLAMF7), and tumor necrosis factor receptor superfamily member 17 (TNFRSF17) were expressed specifically in plasma cells.
For six diseases, the external validity of the protein models could be established in the EPIC-Norfolk study. For four out of the five proteins, which were able to predict more than ten diseases, age was the main correlate.
Comparatively, smoking status was the main correlate for chemokine (C-X-C motif) ligand 17 (CXCL17). Nevertheless, incorporating protein data improved the diagnostic predictability compared to conventional risk factors.
Proteins that were solely and strongly predictive of only one disease were also identified, which included TNFRSF13B for monoclonal gammopathy of undetermined significance (MGUS) and TNFRSF17 or B-cell maturation antigen for MM. In sensitivity analyses, incorporating additional proteins did not necessarily improve model performance; however, including specific biomarkers improved prediction for selected diseases.
Conclusions
Sparse plasma protein signatures can improve prediction as compared to standard clinical assays for common and rare diseases. Nevertheless, future studies are needed to validate these findings in ethnically diverse populations and different geographical regions. For rarer diseases, larger sample sizes are required to estimate detection rates with precision.
Current proteomic platforms provide relative quantification protein assays; however, clinical translation will need further development and validation of absolute quantification protein assays.
Plasma proteins appear to be better for predicting diseases belonging to certain clinical specialties. Therefore, the prediction of other diseases will need to be based on different clinical information.
Journal reference:
- Carrasco-Zanini, J., Pietzner, M., Davitte, J., et al. (2024) Proteomic signatures improve risk prediction for common and rare diseases. Nature Medicine; 1-10. doi:10.1038/s41591-024-03142-z