In a recent study published in the journal Nature Communications, researchers in the United States of America analyzed 23 wildlife species for exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). They investigated the effect of human activity and urbanization on seropositivity. They found the presence of SARS-CoV-2 ribonucleic acid (RNA) in six wildlife species and observed higher seroprevalence in areas with high human activity. Further, they identified seven new human-to-animal transmission events involving the Omicron variant of the virus.
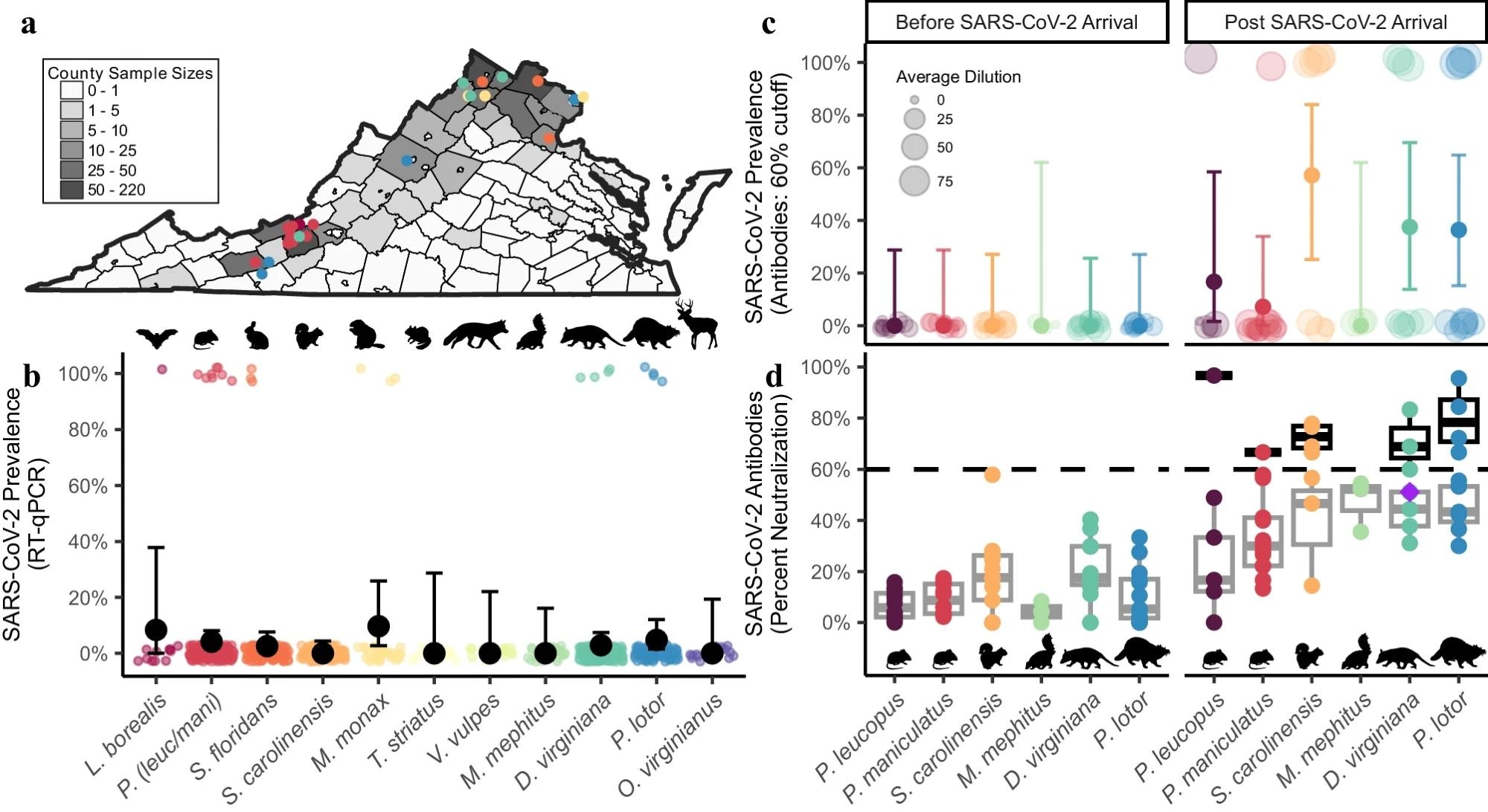 SARS-CoV-2 RNA and neutralizing antibody prevalence in wildlife communities. Study: Widespread exposure to SARS-CoV-2 in wildlife communities
SARS-CoV-2 RNA and neutralizing antibody prevalence in wildlife communities. Study: Widespread exposure to SARS-CoV-2 in wildlife communities
Background
SARS-CoV-2, responsible for COVID-19, has infected over 775 million people and caused over seven million deaths worldwide. As the virus becomes endemic, more virulent variants pose a significant threat. The pandemic has raised concerns about SARS-CoV-2 infecting wildlife and establishing a sylvatic cycle, potentially creating new variants. While transmission to animals in captivity has been documented previously, detection in free-ranging wild animals has been limited mainly to white-tailed deer, feral mink, and Eurasian river otters. Experimental infections suggest that many wildlife species could host the virus, but natural infections remain underexplored.
Numerous variants have emerged since 2019, some increasing infectivity and potentially affecting wildlife. White-tailed deer have shown human-derived and unique viral lineages, suggesting minimal adaptation is needed for deer-to-deer transmission. If SARS-CoV-2 gets established in wildlife, it could lead to novel mutations, potentially increasing virulence, transmissibility, or immune escape. The virus's adaptation to a wide range of hosts could make its evolution more unpredictable, complicating vaccine development and posing new challenges for human health. In the present study, researchers examined SARS-CoV-2 exposure in various species of wildlife across Virginia and Washington D.C. and assessed the impact of urbanization on seropositivity.
About the study
SARS-CoV-2 RNA was extracted from nasopharyngeal/oropharyngeal samples of 789 individual animals from 23 different species. The presence of viral RNA was detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Serum samples from six mammal species (Virginia opossum, raccoon, Eastern gray squirrel, white-footed mouse, deer mouse, and one unspecified species) were collected in 2022 and tested for SARS-CoV-2 antibodies using a 60% neutralization cutoff. Serum samples from pre-2020 and post-2020 eras were compared to assess the differences in neutralization. Additionally, the relationship between urbanization (measured by imperviousness) and wildlife seroprevalence was studied. Human visit data were used to assess the impact of human activity on seroprevalence.
SARS-CoV-2 sequences were obtained from nasal and oropharyngeal swabs using whole genome sequencing. Pango lineages were determined for 12 RNA-positive samples and were compared with human sequences to identify potential recent introductions into wildlife. Unique mutations in the spike (S) protein were studied using molecular modeling and free energy calculations to understand their impact on protein binding and viral infectivity.
Results and discussion
SARS-CoV-2 RNA was detected in nasopharyngeal samples of six of the total species. Out of 789 samples tested, 23 unique individuals were found to be positive. The detections included eight deer mice, four Virginia opossums, four raccoons, three Eastern cottontail rabbits, three groundhogs, and one Eastern red bat. Interestingly, field-collected samples showed a higher positivity rate (4.04%) compared to those from wildlife rehabilitation centers (2.24%).
Furthermore, SARS-CoV-2 antibodies were detected in five of the six species (Virginia opossum, raccoon, Eastern gray squirrel, white-footed mouse, deer mouse) sampled in 2022. Post-2020 samples showed significantly higher neutralization values than pre-2020 samples (p<0.001). Four of the samples were found to exceed an 80% neutralization cutoff.
Additionally, urbanization showed a positive association with wildlife seroprevalence, with the highest antibody detection (80%) at a state park with high human visitation. Human presence was found to be positively associated with seroprevalence, consistent across different neutralization cutoffs.
Sequences from wildlife included Omicron sub-lineages BA.2.10.1 and various XBB* lineages. Notably, the lineages matched those circulating in humans at the time, indicating recent human-to-animal transmissions. The unique E471V mutation in the receptor-binding domain of the S protein was found to improve binding to human angiotensin-converting enzyme 2 (ACE2) receptors compared to the standard BA.2 lineage. The researchers suggest that the unique G798D mutation in the S2 subunit may affect viral entry by affecting glycosylation and structural stability.
Conclusion
In conclusion, the study significantly expanded the known range of wild hosts exposed to SARS-CoV-2, identifying nine species, including six newly documented ones, with the virus. Species such as mice, rabbits, and opossums which tested positive, show traits that may make them more suitable for establishing a SARS-CoV-2 wildlife reservoir. These traits include fast-paced life histories with early and frequent reproduction and a history of being reservoirs for other respiratory viruses.
The findings highlight the need for ongoing broad surveillance and detailed ecological research to fully understand the role of wildlife communities in the transmission and evolution of SARS-CoV-2. In the future, the potential for these species to contribute to the virus's persistence and mutation in wildlife needs to be investigated, as it may have implications for public health strategies.